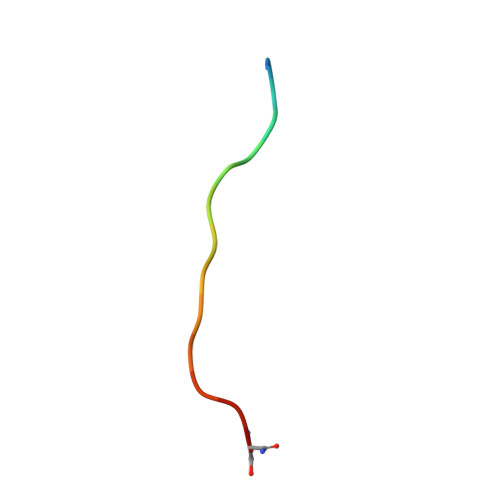
The neuropeptide galanin adopts an irregular secondary structure.
Wilkinson, R.E., Kraichely, K.N., Hendy, C.M., Buchanan, L.E., Parnham, S., Giuliano, M.W.(2022) Biochem Biophys Res Commun 626: 121-128
- PubMed: 35994823
- DOI: https://doi.org/10.1016/j.bbrc.2022.08.032
- Primary Citation of Related Structures:
8DHZ, 8DJ4 - PubMed Abstract:
Human galanin is a 30-residue neuropeptide targeted for development of analgesics, antidepressants, and anticonvulsants. While previous work from our group and others has already produced significant insights into galanin's N-terminal region, no extant structures of galanin in databases include its full-length sequence and the function of its C-terminus remains ambiguous. We report the NMR solution structure of full-length human galanin C-terminal amide, determined from 2D 1 H- 1 H COSY, TOCSY, and ROESY NMR data. Galanin adopts an irregular helical structure across its N-terminus, likely the average of several coiling states. We present the NMR structure of a peptide encompassing the C-terminus of galanin as a stand-alone fragment. The C-terminus of full-length galanin appears to indirectly assist the intramolecular association of hydrophobic sidechains within its N-terminus, remotely rigidifying their position when compared to previously studied N-terminal galanin fragments. By contrast, there is flexibility in the C-terminus of galanin, characterized by two i to i + 2 hydrogen-bonded turns within an otherwise dynamic backbone. The C-terminal portion of the peptide renders it soluble, and plays a hitherto undescribed biophysical role in pre-organizing the galanin receptor binding epitope. We speculate that hydrophilic microdomains of signaling peptides, hormones, and perhaps intrinsically disordered proteins may also function similarly.
- Department of Chemistry and Biochemistry, College of Charleston, Charleston, SC, 29424, USA.
Organizational Affiliation: