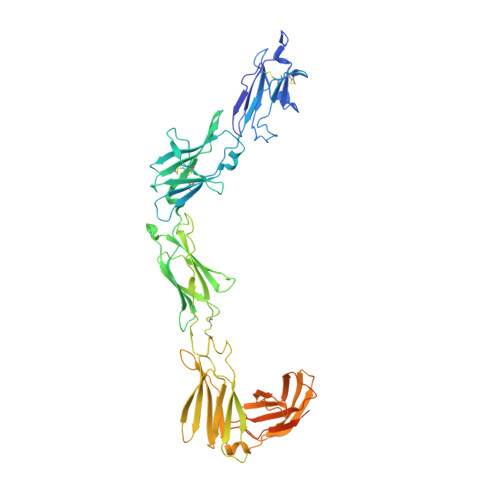
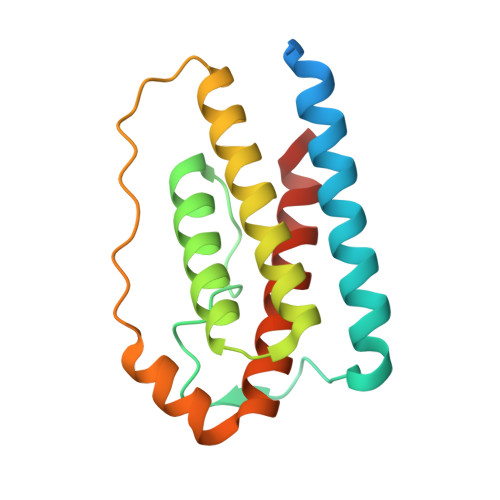
Structural insights into the mechanism of leptin receptor activation.
Saxton, R.A., Caveney, N.A., Moya-Garzon, M.D., Householder, K.D., Rodriguez, G.E., Burdsall, K.A., Long, J.Z., Garcia, K.C.(2023) Nat Commun 14: 1797-1797
- PubMed: 37002197
- DOI: https://doi.org/10.1038/s41467-023-37169-6
- Primary Citation of Related Structures:
8DH8, 8DH9, 8DHA - PubMed Abstract:
Leptin is an adipocyte-derived protein hormone that promotes satiety and energy homeostasis by activating the leptin receptor (LepR)-STAT3 signaling axis in a subset of hypothalamic neurons. Leptin signaling is dysregulated in obesity, however, where appetite remains elevated despite high levels of circulating leptin. To gain insight into the mechanism of leptin receptor activation, here we determine the structure of a stabilized leptin-bound LepR signaling complex using single particle cryo-EM. The structure reveals an asymmetric architecture in which a single leptin induces LepR dimerization via two distinct receptor-binding sites. Analysis of the leptin-LepR binding interfaces reveals the molecular basis for human obesity-associated mutations. Structure-based design of leptin variants that destabilize the asymmetric LepR dimer yield both partial and biased agonists that partially suppress STAT3 activation in the presence of wild-type leptin and decouple activation of STAT3 from LepR negative regulators. Together, these results reveal the structural basis for LepR activation and provide insights into the differential plasticity of signaling pathways downstream of LepR.
- Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, 94305, USA. rsaxton@berkeley.edu.
Organizational Affiliation: