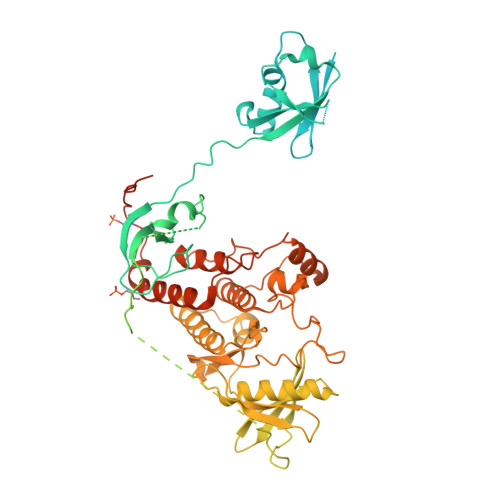
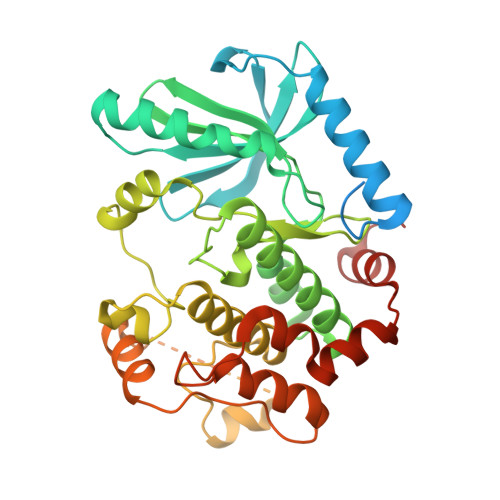
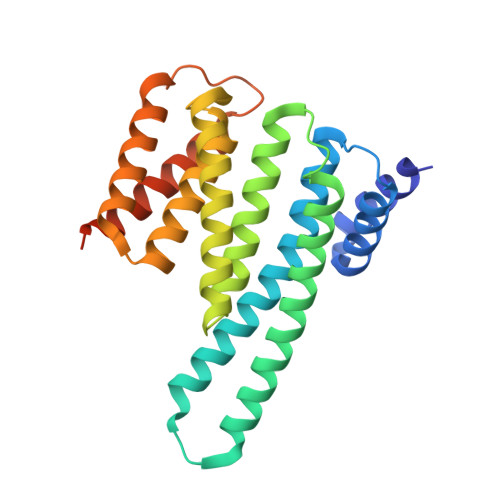
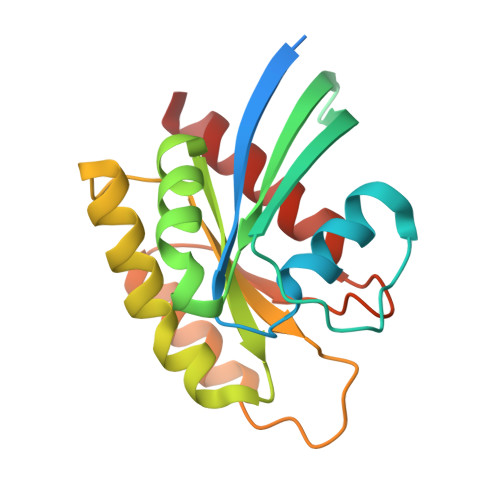
Cryo-EM structure of a RAS/RAF recruitment complex.
Park, E., Rawson, S., Schmoker, A., Kim, B.W., Oh, S., Song, K., Jeon, H., Eck, M.J.(2023) Nat Commun 14: 4580-4580
- PubMed: 37516774
- DOI: https://doi.org/10.1038/s41467-023-40299-6
- Primary Citation of Related Structures:
8DGS, 8DGT - PubMed Abstract:
RAF-family kinases are activated by recruitment to the plasma membrane by GTP-bound RAS, whereupon they initiate signaling through the MAP kinase cascade. Prior structural studies of KRAS with RAF have focused on the isolated RAS-binding and cysteine-rich domains of RAF (RBD and CRD, respectively), which interact directly with RAS. Here we describe cryo-EM structures of a KRAS bound to intact BRAF in an autoinhibited state with MEK1 and a 14-3-3 dimer. Analysis of this KRAS/BRAF/MEK1/14-3-3 complex reveals KRAS bound to the RAS-binding domain of BRAF, captured in two orientations. Core autoinhibitory interactions in the complex are unperturbed by binding of KRAS and in vitro activation studies confirm that KRAS binding is insufficient to activate BRAF, absent membrane recruitment. These structures illustrate the separability of binding and activation of BRAF by RAS and suggest stabilization of this pre-activation intermediate as an alternative therapeutic strategy to blocking binding of KRAS.
- Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA, 02215, USA.
Organizational Affiliation: