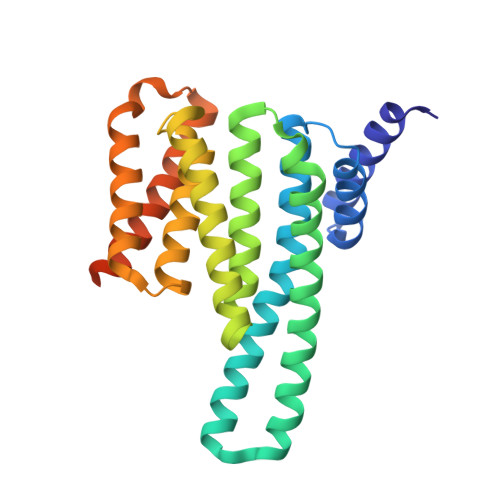
Structural mapping of PEAK pseudokinase interactions identifies 14-3-3 as a molecular switch for PEAK3 signaling.
Roy, M.J., Surudoi, M.G., Kropp, A., Hou, J., Dai, W., Hardy, J.M., Liang, L.Y., Cotton, T.R., Lechtenberg, B.C., Dite, T.A., Ma, X., Daly, R.J., Patel, O., Lucet, I.S.(2023) Nat Commun 14: 3542-3542
- PubMed: 37336884
- DOI: https://doi.org/10.1038/s41467-023-38869-9
- Primary Citation of Related Structures:
8DGM, 8DGN, 8DGO, 8DGP - PubMed Abstract:
PEAK pseudokinases regulate cell migration, invasion and proliferation by recruiting key signaling proteins to the cytoskeleton. Despite lacking catalytic activity, alteration in their expression level is associated with several aggressive cancers. Here, we elucidate the molecular details of key PEAK signaling interactions with the adapter proteins CrkII and Grb2 and the scaffold protein 14-3-3. Our findings rationalize why the dimerization of PEAK proteins has a crucial function in signal transduction and provide biophysical and structural data to unravel binding specificity within the PEAK interactome. We identify a conserved high affinity 14-3-3 motif on PEAK3 and demonstrate its role as a molecular switch to regulate CrkII binding and signaling via Grb2. Together, our studies provide a detailed structural snapshot of PEAK interaction networks and further elucidate how PEAK proteins, especially PEAK3, act as dynamic scaffolds that exploit adapter proteins to control signal transduction in cell growth/motility and cancer.
- The Walter and Eliza Hall Institute of Medical Research, Parkville, VIC, 3052, Australia. roy@wehi.edu.au.
Organizational Affiliation: