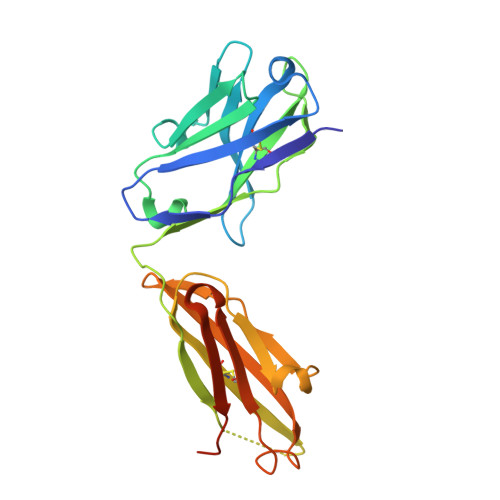
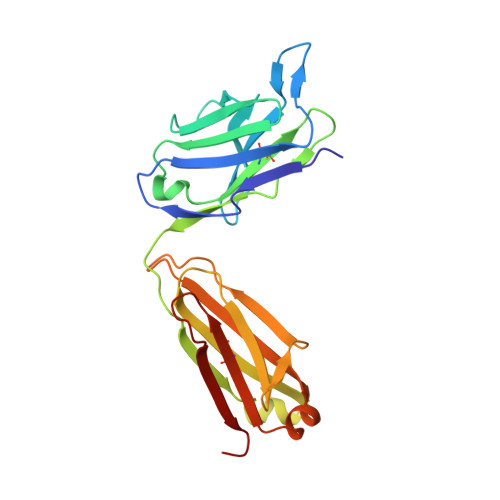
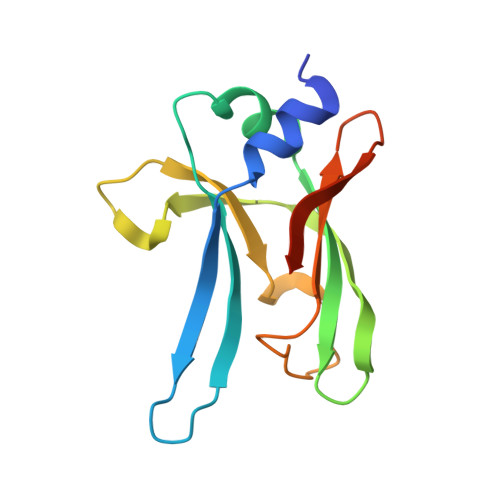
Structural Basis for Binding of Neutralizing Antibodies to Clostridioides difficile Binary Toxin.
Goldsmith, J.A., Dewar, V., Hermand, P., Blais, N., McLellan, J.S.(2023) J Bacteriol 205: e0045622-e0045622
- PubMed: 36951574
- DOI: https://doi.org/10.1128/jb.00456-22
- Primary Citation of Related Structures:
8DCM, 8DCN - PubMed Abstract:
Clostridioides difficile is a Gram-positive opportunistic human pathogen that causes 15,000 deaths annually in the United States, prompting a need for vaccine development. In addition to the important toxins TcdA and TcdB, binary toxin (CDT) plays a significant role in the pathogenesis of certain C. difficile ribotypes by catalyzing the ADP-ribosylation of actin in host cells. However, the mechanisms of CDT neutralization by antibodies have not been studied, limiting our understanding of key epitopes for CDT antigen design. Therefore, we isolated neutralizing monoclonal antibodies against CDT and characterized their mechanisms of neutralization structurally and biochemically. Here, 2.5-Å and 2.6-Å resolution X-ray crystal structures of the antibodies BINTOXB/22 and BINTOXB/9, respectively, in complex with CDTb-the CDT subunit that forms a heptameric pore for the delivery of toxic CDTa enzyme into the host cytosol-showed that both antibodies sterically clash with adjacent protomers in the assembled heptamer. Assessment of trypsin-induced oligomerization of the purified CDTb protoxin in vitro showed that BINTOXB/22 and BINTOXB/9 prevented the assembly of di-heptamers upon prodomain cleavage. This work suggests that the CDT oligomerization process can be effectively targeted by antibodies, which will aid in the development of C. difficile vaccines and therapeutics. IMPORTANCE Clostridioides difficile strains associated with worse clinical outcomes have been found to secrete a toxin called CDT (or binary toxin). As blocking the function of this toxin could help mitigate C. difficile infections, we sought to determine the molecular basis for the inhibition of CDT by monoclonal antibodies. We isolated monoclonal antibodies targeting the B-component of CDT (CDTb) and selected two with neutralizing activity for detailed structural and biochemical characterization. High-resolution crystal structures of each antibody bound to CDTb showed that their presence would preclude the assembly of a CDTb oligomer required for activity. Oligomerization of CDTb in vitro was shown to be blocked in the presence of the neutralizing antibodies, but not a control antibody.
- Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas, USA.
Organizational Affiliation: