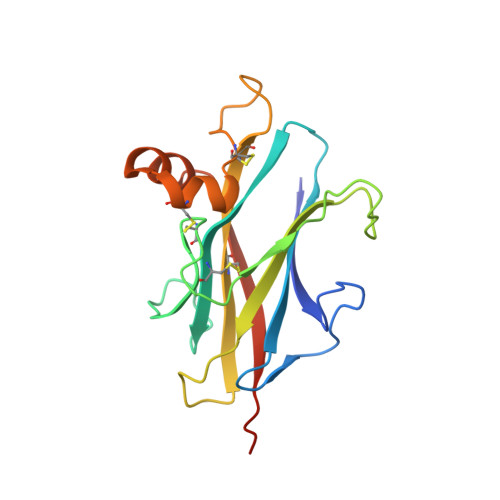
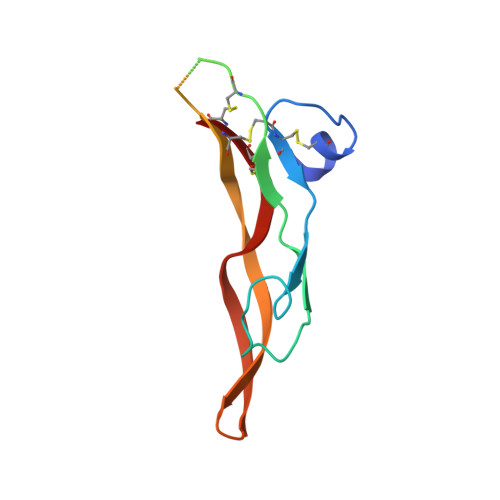
Structures of TGF-beta with betaglycan and signaling receptors reveal mechanisms of complex assembly and signaling.
Wieteska, L., Taylor, A.B., Punch, E., Coleman, J.A., Conway, I.O., Lin, Y.F., Byeon, C.H., Hinck, C.S., Krzysiak, T., Ishima, R., Lopez-Casillas, F., Cherepanov, P., Bernard, D.J., Hill, C.S., Hinck, A.P.(2025) Nat Commun 16: 1778-1778
- PubMed: 40011426
- DOI: https://doi.org/10.1038/s41467-025-56796-9
- Primary Citation of Related Structures:
8DC0, 9B9F, 9FDY, 9FK5, 9FKP - PubMed Abstract:
Betaglycan (BG) is a transmembrane co-receptor of the transforming growth factor-β (TGF-β) family of signaling ligands. It is essential for embryonic development, tissue homeostasis and fertility in adults. It functions by enabling binding of the three TGF-β isoforms to their signaling receptors and is additionally required for inhibin A (InhA) activity. Despite its requirement for the functions of TGF-βs and InhA in vivo, structural information explaining BG ligand selectivity and its mechanism of action is lacking. Here, we determine the structure of TGF-β bound both to BG and the signaling receptors, TGFBR1 and TGFBR2. We identify key regions responsible for ligand engagement, which has revealed binding interfaces that differ from those described for the closely related co-receptor of the TGF-β family, endoglin, thus demonstrating remarkable evolutionary adaptation to enable ligand selectivity. Finally, we provide a structural explanation for the hand-off mechanism underlying TGF-β signal potentiation.
- Department of Structural Biology, University of Pittsburgh, Pittsburgh, PA, USA.
Organizational Affiliation: