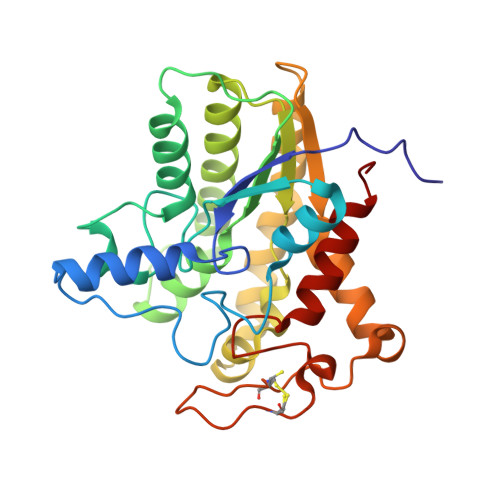
Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
Pham, V.D., Couture, M., Lortie, L.-A., Picard, M.-E., Charette, S., Levesque, R., Shi, R.To be published.