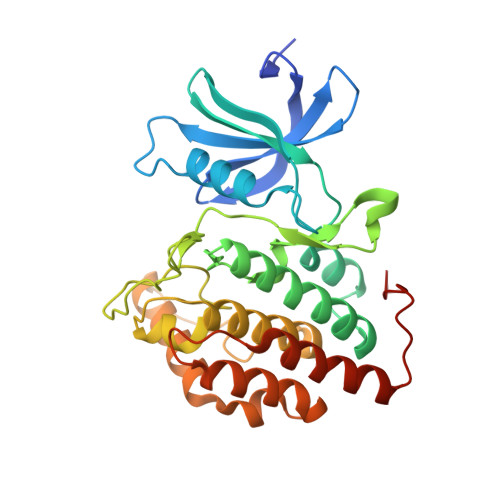
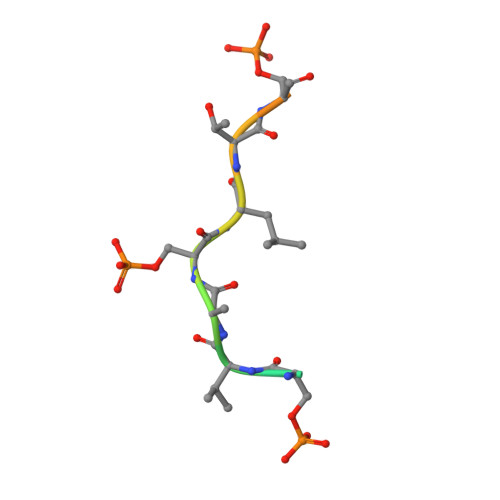
PERIOD phosphorylation leads to feedback inhibition of CK1 activity to control circadian period.
Philpott, J.M., Freeberg, A.M., Park, J., Lee, K., Ricci, C.G., Hunt, S.R., Narasimamurthy, R., Segal, D.H., Robles, R., Cai, Y., Tripathi, S., McCammon, J.A., Virshup, D.M., Chiu, J.C., Lee, C., Partch, C.L.(2023) Mol Cell 83: 1677-1692.e8
- PubMed: 37207626
- DOI: https://doi.org/10.1016/j.molcel.2023.04.019
- Primary Citation of Related Structures:
8D7M, 8D7N, 8D7O, 8D7P - PubMed Abstract:
PERIOD (PER) and Casein Kinase 1δ regulate circadian rhythms through a phosphoswitch that controls PER stability and repressive activity in the molecular clock. CK1δ phosphorylation of the familial advanced sleep phase (FASP) serine cluster embedded within the Casein Kinase 1 binding domain (CK1BD) of mammalian PER1/2 inhibits its activity on phosphodegrons to stabilize PER and extend circadian period. Here, we show that the phosphorylated FASP region (pFASP) of PER2 directly interacts with and inhibits CK1δ. Co-crystal structures in conjunction with molecular dynamics simulations reveal how pFASP phosphoserines dock into conserved anion binding sites near the active site of CK1δ. Limiting phosphorylation of the FASP serine cluster reduces product inhibition, decreasing PER2 stability and shortening circadian period in human cells. We found that Drosophila PER also regulates CK1δ via feedback inhibition through the phosphorylated PER-Short domain, revealing a conserved mechanism by which PER phosphorylation near the CK1BD regulates CK1 kinase activity.
- Department of Chemistry and Biochemistry, University of California, Santa Cruz, Santa Cruz, CA 95064, USA.
Organizational Affiliation: