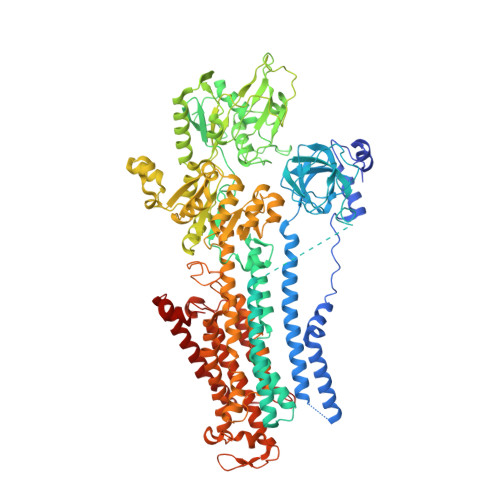
Structural basis for gating mechanism of the human sodium-potassium pump.
Nguyen, P.T., Deisl, C., Fine, M., Tippetts, T.S., Uchikawa, E., Bai, X.C., Levine, B.(2022) Nat Commun 13: 5293-5293
- PubMed: 36075933
- DOI: https://doi.org/10.1038/s41467-022-32990-x
- Primary Citation of Related Structures:
8D3U, 8D3V, 8D3W, 8D3X, 8D3Y - PubMed Abstract:
P2-type ATPase sodium-potassium pumps (Na + /K + -ATPases) are ion-transporting enzymes that use ATP to transport Na + and K + on opposite sides of the lipid bilayer against their electrochemical gradients to maintain ion concentration gradients across the membranes in all animal cells. Despite the available molecular architecture of the Na + /K + -ATPases, a complete molecular mechanism by which the Na + and K + ions access into and are released from the pump remains unknown. Here we report five cryo-electron microscopy (cryo-EM) structures of the human alpha3 Na + /K + -ATPase in its cytoplasmic side-open (E1), ATP-bound cytoplasmic side-open (E1•ATP), ADP-AlF 4 - trapped Na + -occluded (E1•P-ADP), BeF 3 - trapped exoplasmic side-open (E2P) and MgF 4 2- trapped K + -occluded (E2•P i ) states. Our work reveals the atomically resolved structural detail of the cytoplasmic gating mechanism of the Na + /K + -ATPase.
- Howard Hughes Medical Institute and Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX, USA. phong.nguyen@utsouthwestern.edu.
Organizational Affiliation: