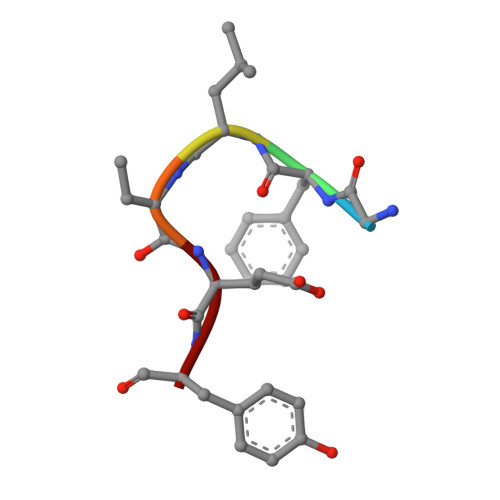
The mechanism of thia-Michael addition catalyzed by LanC enzymes.
Ongpipattanakul, C., Liu, S., Luo, Y., Nair, S.K., van der Donk, W.A.(2023) Proc Natl Acad Sci U S A 120: e2217523120-e2217523120
- PubMed: 36634136
- DOI: https://doi.org/10.1073/pnas.2217523120
- Primary Citation of Related Structures:
8CZK, 8CZL, 8D0V, 8D19 - PubMed Abstract:
In both eukarya and bacteria, the addition of Cys to dehydroalanine (Dha) and dehydrobutyrine (Dhb) occurs in various biological processes. In bacteria, intramolecular thia-Michael addition catalyzed by lanthipeptide cyclases (LanC) proteins or protein domains gives rise to a class of natural products called lanthipeptides. In eukarya, dehydroamino acids in signaling proteins are introduced by effector proteins produced by pathogens like Salmonella to dysregulate host defense mechanisms. A eukaryotic LanC-like (LanCL) enzyme catalyzes the addition of Cys in glutathione to Dha/Dhb to protect the cellular proteome from unwanted chemical and biological activity. To date, the mechanism of the enzyme-catalyzed thia-Michael addition has remained elusive. We report here the crystal structures of the human LanCL1 enzyme complexed with different ligands, including the product of thia-Michael addition of glutathione to a Dhb-containing peptide that represents the activation loop of Erk. The structures show that a zinc ion activates the Cys thiolate for nucleophilic attack and that a conserved His is poised to protonate the enolate intermediate to achieve a net anti- addition. A second His hydrogen bonds to the carbonyl oxygen of the former Dhb and may stabilize the negative charge that builds up on this oxygen atom in the enolate intermediate. Surprisingly, the latter His is not conserved in orthologous enzymes that catalyze thia-Michael addition to Dha/Dhb. Eukaryotic LanCLs contain a His, whereas bacterial stand-alone LanCs have a Tyr residue, and LanM enzymes that have LanC-like domains have a Lys, Asn, or His residue. Mutational and binding studies support the importance of these residues for catalysis.
- Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801.
Organizational Affiliation: