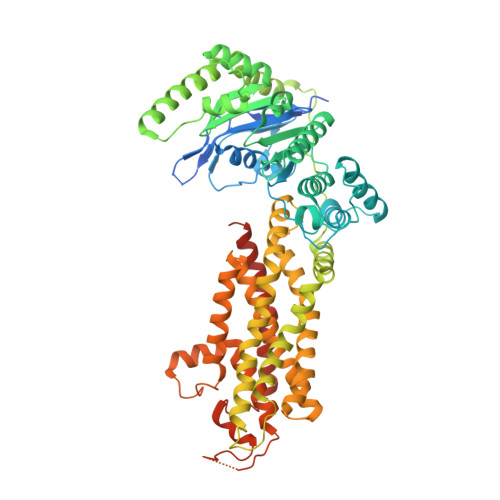
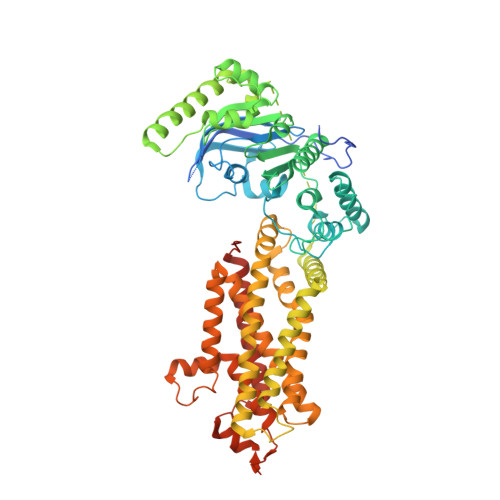
Structural Analysis of Cholesterol Binding and Sterol Selectivity by ABCG5/G8.
Farhat, D., Rezaei, F., Ristovski, M., Yang, Y., Stancescu, A., Dzimkova, L., Samnani, S., Couture, J.F., Lee, J.Y.(2022) J Mol Biology 434: 167795-167795
- PubMed: 35988751
- DOI: https://doi.org/10.1016/j.jmb.2022.167795
- Primary Citation of Related Structures:
8CUB - PubMed Abstract:
The ATP-binding cassette (ABC) sterol transporters are responsible for maintaining cholesterol homeostasis in mammals by participating in reverse cholesterol transport (RCT) or transintestinal cholesterol efflux (TICE). The heterodimeric ABCG5/G8 carries out selective sterol excretion, preventing the abnormal accumulation of plant sterols in human bodies, while homodimeric ABCG1 contributes to the biogenesis and metabolism of high-density lipoproteins. A sterol-binding site on ABCG5/G8 was proposed at the interface of the transmembrane domain and the core of lipid bilayers. In this study, we have determined the crystal structure of ABCG5/G8 in a cholesterol-bound state. The structure combined with amino acid sequence analysis shows that in the proximity of the sterol-binding site, a highly conserved phenylalanine array supports functional implications for ABCG cholesterol/sterol transporters. Lastly, in silico docking analysis of cholesterol and stigmasterol (a plant sterol) suggests sterol-binding selectivity on ABCG5/G8, but not ABCG1. Together, our results provide a structural basis for cholesterol binding on ABCG5/G8 and the sterol selectivity by ABCG transporters.
- Department of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Ottawa, Ontario, Canada. Electronic address: https://twitter.com/FarhDanny.
Organizational Affiliation: