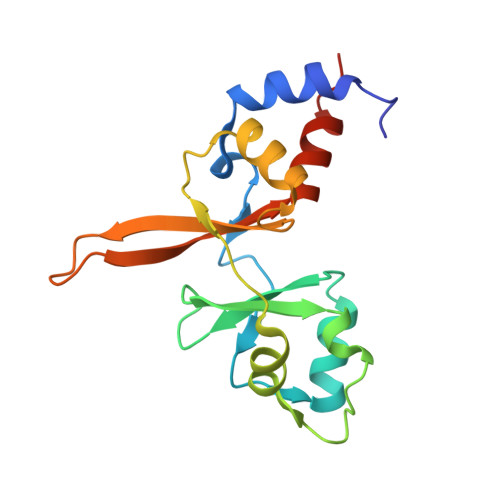
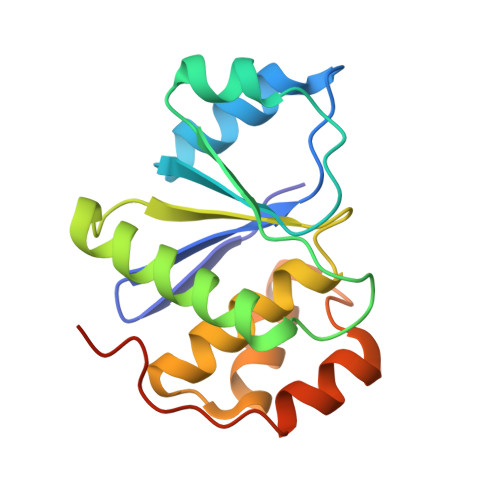
Burst kinetics and CNNM binding are evolutionarily conserved properties of phosphatases of regenerating liver.
Fakih, R., Goldstein, R.H., Kozlov, G., Gehring, K.(2023) J Biological Chem 299: 103055-103055
- PubMed: 36822330
- DOI: https://doi.org/10.1016/j.jbc.2023.103055
- Primary Citation of Related Structures:
8CT8 - PubMed Abstract:
Phosphatases of regenerating liver (PRL or PTP4A) are a family of enigmatic protein phosphatases implicated in cell growth and metabolism. Despite their relevance in metastatic cancer, much remains unknown about the PRL family. They act as pseudophosphatases to regulate the CNNM family of magnesium transporters yet also have enzymatic activity on unknown substrates. In mammals, PRLs are mostly found trapped in an intermediate state that regulates their pseudophosphatase activity. Phosphocysteine, which is formed as an intermediate in the phosphatase catalytic cycle, is inefficiently hydrolyzed leading to burst enzyme kinetics and turnover numbers of less than one per hour. In flies, PRLs have recently been shown to have neuroprotective and neurodevelopmental roles raising the question whether they act as phosphatases, pseudophosphatases, or both. Here, we characterize the evolutionary development of PRLs and ask whether their unique structural and functional properties are conserved. We purified recombinant PRL proteins from 15 phylogenetically diverse organisms and characterized their catalytic activities and ability to bind CNNM proteins. We observed PRLs from humans to amoebae form a stable phosphocysteine intermediate and exhibit burst kinetics. Isothermal titration calorimetry experiments confirmed that the PRL-CNNM interaction is broadly conserved with nanomolar affinity in vertebrates. Lastly, we determined the crystal structure of the Drosophila melanogaster PRL-CNNM complex and identified mutants that specifically impair either phosphatase activity or CNNM binding. Our results reveal the unique properties of PRLs are conserved throughout the animal kingdom and open the door to using model organisms to dissect PRL function in cell signaling.
- Department of Biochemistry, Centre for Structural Biology, McGill University, Montreal, Quebec, Canada.
Organizational Affiliation: