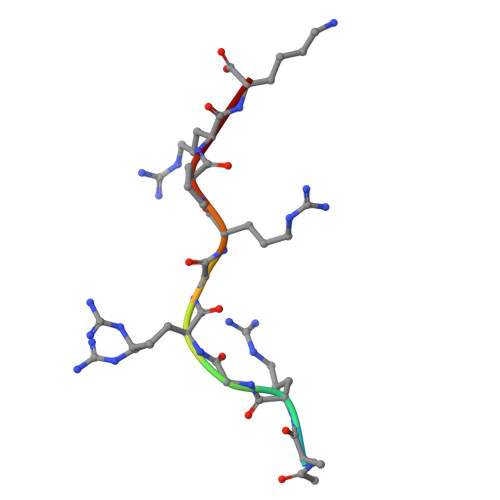
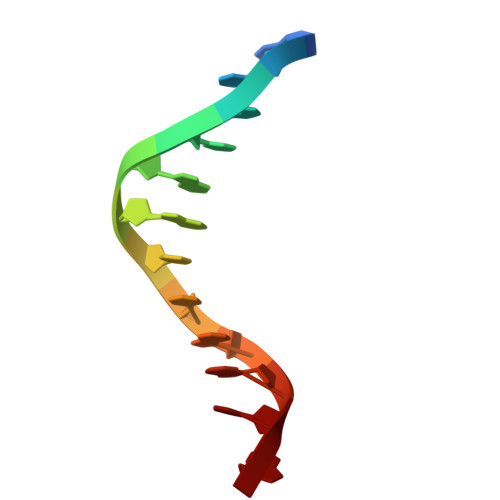
Crystal structure of the HMGA AT-hook 1 domain bound to the minor groove of AT-rich DNA and inhibition by antikinetoplastid drugs.
Nue-Martinez, J.J., Maturana, M., Lagartera, L., Rodriguez-Gutierrez, J.A., Boer, R., Campos, J.L., Saperas, N., Dardonville, C.(2024) Sci Rep 14: 26173-26173
- PubMed: 39478017
- DOI: https://doi.org/10.1038/s41598-024-77522-3
- Primary Citation of Related Structures:
8CPG - PubMed Abstract:
High mobility group (HMG) proteins are intrinsically disordered nuclear non-histone chromosomal proteins that play an essential role in many biological processes by regulating the expression of numerous genes in eukaryote cells. HMGA proteins contain three DNA binding motifs, the "AT-hooks", that bind preferentially to AT-rich sequences in the minor groove of B-form DNA. Understanding the interactions of AT-hook domains with DNA is very relevant from a medical point of view because HMGA proteins are involved in different conditions including cancer and parasitic diseases. We present here the first crystal structure (1.40 Å resolution) of the HMGA AT-hook 1 domain, bound to the minor groove of AT-rich DNA. In contrast to AT-hook 3 which bends DNA and shows a larger minor groove widening, AT-hook 1 binds neighbouring DNA molecules and displays moderate widening of DNA upon binding. The binding affinity and thermodynamics of binding were studied in solution with surface plasmon resonance (SPR)-biosensor and isothermal titration calorimetry (ITC) experiments. AT-hook 1 forms an entropy-driven 2:1 complex with (TTAA) 2 -containing DNA with relatively slow kinetics of association/dissociation. We show that N-phenylbenzamide-derived antikinetoplastid compounds (1-3) bind strongly and specifically to the minor groove of AT-DNA and compete with AT-hook 1 for binding. The central core of the molecule is the basis for the observed sequence selectivity of these compounds. These findings provide clues regarding a possible mode of action of DNA minor groove binding compounds that are relevant to major neglected tropical diseases such as leishmaniasis and trypanosomiasis.
- Instituto de Química Médica, IQM-CSIC, Madrid, 28006, Spain.
Organizational Affiliation: