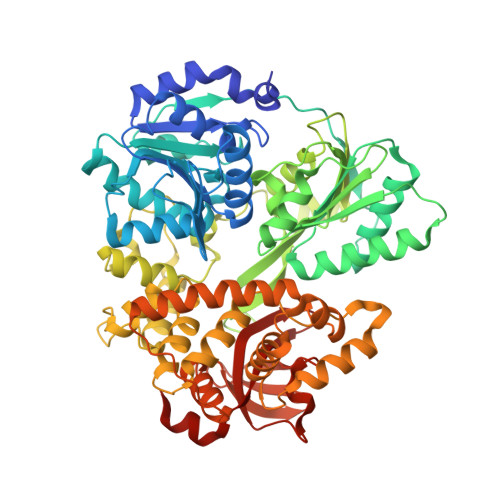
Crystal structure of Prp16 in complex with ADP.
Garbers, T.B., Enders, M., Neumann, P., Ficner, R.(2023) Acta Crystallogr F Struct Biol Commun 79: 200-207
- PubMed: 37548918
- DOI: https://doi.org/10.1107/S2053230X23005721
- Primary Citation of Related Structures:
8CNT - PubMed Abstract:
DEAH-box helicases play a crucial role in pre-mRNA splicing as they are responsible for major rearrangements of the spliceosome and are involved in various quality-ensuring steps. Prp16 is the driving force during spliceosomal catalysis, remodeling the C state into the C* state. Here, the first crystal structure of Prp16 from Chaetomium thermophilum in complex with ADP is reported at 1.9 Å resolution. Comparison with the other spliceosomal DEAH-box helicases Prp2, Prp22 and Prp43 reveals an overall identical domain architecture. The β-hairpin, which is a structural element of the RecA2 domain, exhibits a unique position, punctuating its flexibility. Analysis of cryo-EM models of spliceosomal complexes containing Prp16 reveals that these models show Prp16 in its nucleotide-free state, rendering the model presented here the first structure of Prp16 in complex with a nucleotide.
- Department of Molecular Structural Biology, Institute of Microbiology and Genetics, GZMB, Georg-August-University Göttingen, Göttingen, Germany.
Organizational Affiliation: