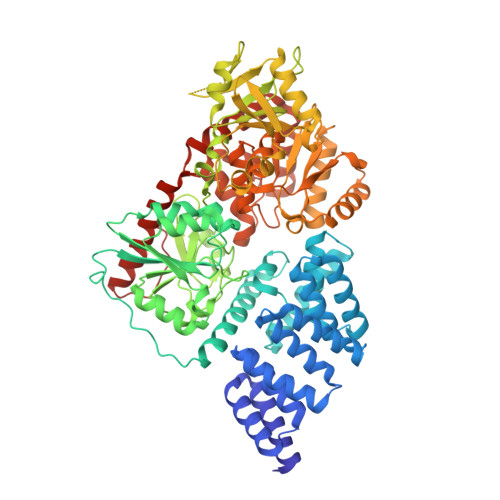
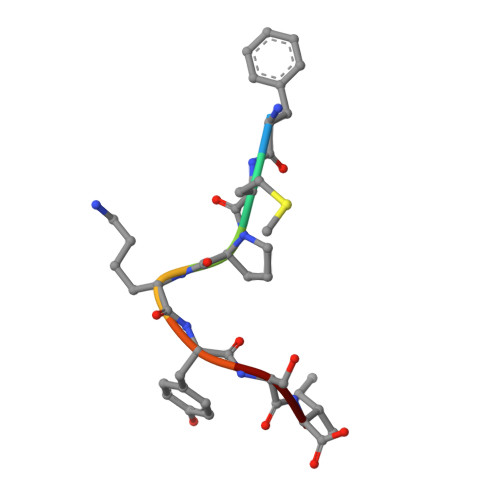
Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase.
Alteen, M.G., Meek, R.W., Kolappan, S., Busmann, J.A., Cao, J., O'Gara, Z., Chou, Y., Derda, R., Davies, G.J., Vocadlo, D.J.(2023) Proc Natl Acad Sci U S A 120: e2303690120-e2303690120
- PubMed: 37819980
- DOI: https://doi.org/10.1073/pnas.2303690120
- Primary Citation of Related Structures:
8CM9 - PubMed Abstract:
The modification of nucleocytoplasmic proteins by O -linked N-acetylglucosamine ( O -GlcNAc) is an important regulator of cell physiology. O -GlcNAc is installed on over a thousand proteins by just one enzyme, O -GlcNAc transferase (OGT). How OGT is regulated is therefore a topic of interest. To gain insight into these questions, we used OGT to perform phage display selection from an unbiased library of ~10 9 peptides of 15 amino acids in length. Following rounds of selection and deep mutational panning, we identified a high-fidelity peptide consensus sequence, [Y/F]-x-P-x-Y-x-[I/M/F], that drives peptide binding to OGT. Peptides containing this sequence bind to OGT in the high nanomolar to low micromolar range and inhibit OGT in a noncompetitive manner with low micromolar potencies. X-ray structural analyses of OGT in complex with a peptide containing this motif surprisingly revealed binding to an exosite proximal to the active site of OGT. This structure defines the detailed molecular basis driving peptide binding and explains the need for specific residues within the sequence motif. Analysis of the human proteome revealed this motif within 52 nuclear and cytoplasmic proteins. Collectively, these data suggest a mode of regulation of OGT by which polypeptides can bind to this exosite to cause allosteric inhibition of OGT through steric occlusion of its active site. We expect that these insights will drive improved understanding of the regulation of OGT within cells and enable the development of new chemical tools to exert fine control over OGT activity.
- Department of Chemistry, Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
Organizational Affiliation: