Molecular Mechanism of Sirtuin 1 Inhibition by Human Immunodeficiency Virus 1 Tat Protein.
Adolph, R.S., Beck, E., Schweimer, K., Di Fonzo, A., Weyand, M., Rosch, P., Wohrl, B.M., Steegborn, C.(2023) Life (Basel) 13
- PubMed: 37109478
- DOI: https://doi.org/10.3390/life13040949
- Primary Citation of Related Structures:
8CCW, 8CCZ - PubMed Abstract:
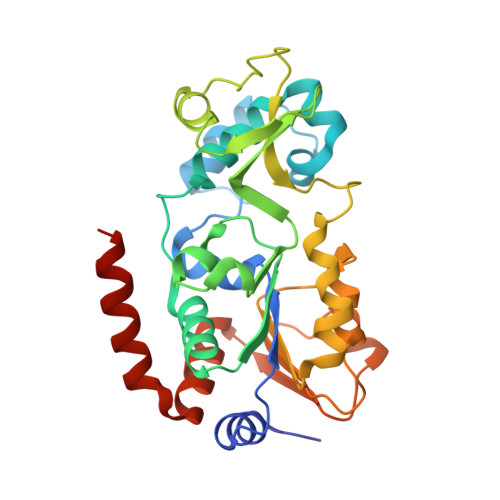
Sirtuins are NAD + -dependent protein lysine deacylases implicated in metabolic regulation and aging-related dysfunctions. The nuclear isoform Sirt1 deacetylates histones and transcription factors and contributes, e.g., to brain and immune cell functions. Upon infection by human immunodeficiency virus 1 (HIV1), Sirt1 deacetylates the viral transactivator of transcription (Tat) protein to promote the expression of the viral genome. Tat, in turn, inhibits Sirt1, leading to the T cell hyperactivation associated with HIV infection. Here, we describe the molecular mechanism of Tat-dependent sirtuin inhibition. Using Tat-derived peptides and recombinant Tat protein, we mapped the inhibitory activity to Tat residues 34-59, comprising Tat core and basic regions and including the Sirt1 deacetylation site Lys50. Tat binds to the sirtuin catalytic core and inhibits Sirt1, Sirt2, and Sirt3 with comparable potencies. Biochemical data and crystal structures of sirtuin complexes with Tat peptides reveal that Tat exploits its intrinsically extended basic region for binding to the sirtuin substrate binding cleft through substrate-like β-strand interactions, supported by charge complementarity. Tat Lys50 is positioned in the sirtuin substrate lysine pocket, although binding and inhibition do not require prior acetylation and rely on subtle differences to the binding of regular substrates. Our results provide mechanistic insights into sirtuin regulation by Tat, improving our understanding of physiological sirtuin regulation and the role of this interaction during HIV1 infection.
- Department of Biochemistry, University of Bayreuth, 95440 Bayreuth, Germany.
Organizational Affiliation: