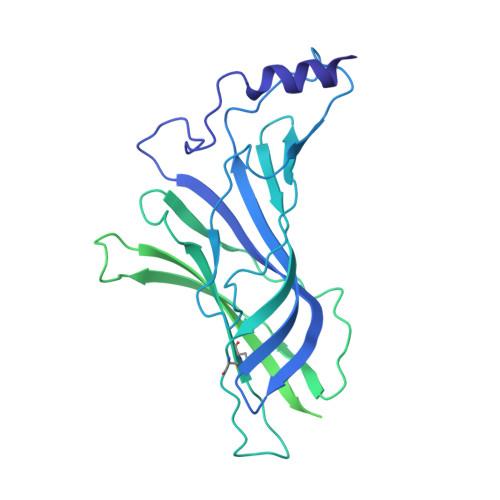
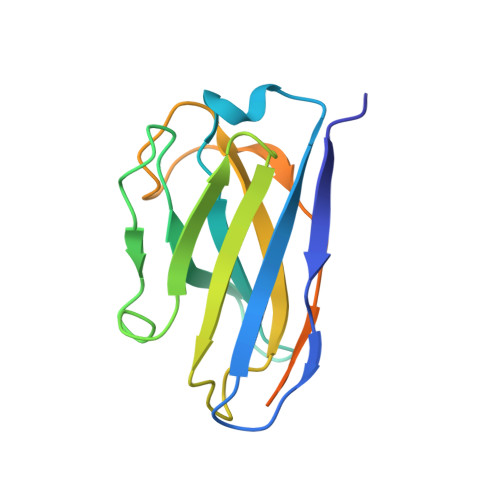
An original potentiating mechanism revealed by the cryo-EM structures of the human alpha 7 nicotinic receptor in complex with nanobodies.
Prevost, M.S., Barilone, N., Dejean de la Batie, G., Pons, S., Ayme, G., England, P., Gielen, M., Bontems, F., Pehau-Arnaudet, G., Maskos, U., Lafaye, P., Corringer, P.J.(2023) Nat Commun 14: 5964-5964
- PubMed: 37749098
- DOI: https://doi.org/10.1038/s41467-023-41734-4
- Primary Citation of Related Structures:
8C9X, 8CAU, 8CE4, 8CI1, 8CI2 - PubMed Abstract:
The human α7 nicotinic receptor is a pentameric channel mediating cellular and neuronal communication. It has attracted considerable interest in designing ligands for the treatment of neurological and psychiatric disorders. To develop a novel class of α7 ligands, we recently generated two nanobodies named E3 and C4, acting as positive allosteric modulator and silent allosteric ligand, respectively. Here, we solved the cryo-electron microscopy structures of the nanobody-receptor complexes. E3 and C4 bind to a common epitope involving two subunits at the apex of the receptor. They form by themselves a symmetric pentameric assembly that extends the extracellular domain. Unlike C4, the binding of E3 drives an agonist-bound conformation of the extracellular domain in the absence of an orthosteric agonist, and mutational analysis shows a key contribution of an N-linked sugar moiety in mediating E3 potentiation. The nanobody E3, by remotely controlling the global allosteric conformation of the receptor, implements an original mechanism of regulation that opens new avenues for drug design.
- Institut Pasteur, Université Paris Cité, CNRS UMR 3571, Channel-Receptors Unit, Paris, France. marie.prevost@pasteur.fr.
Organizational Affiliation: