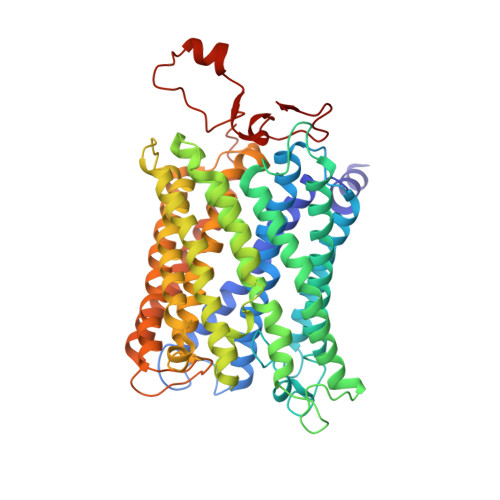
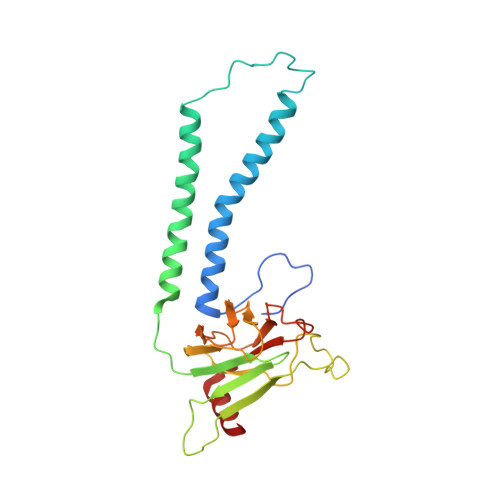
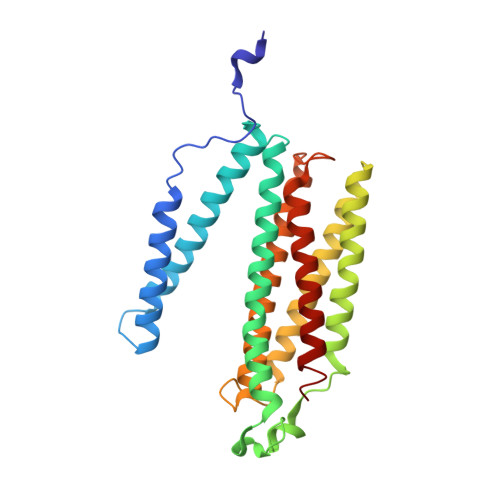
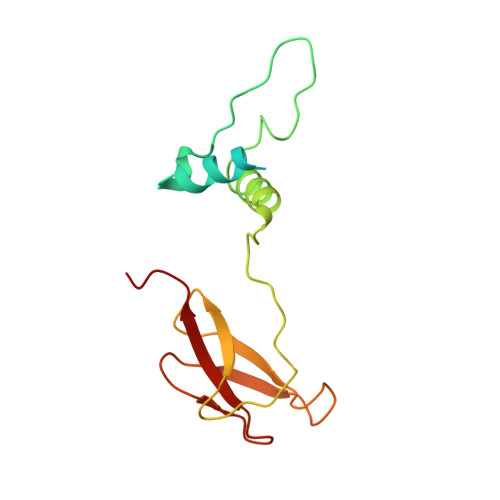
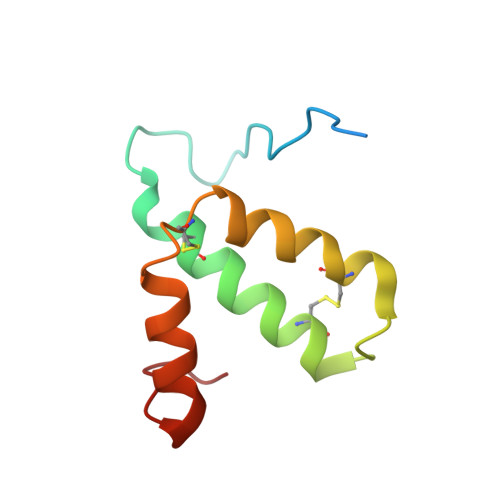
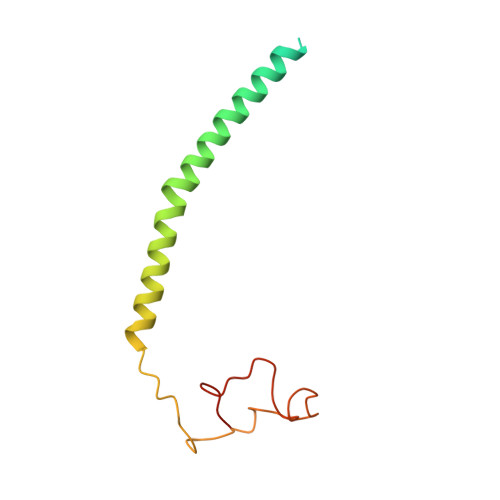
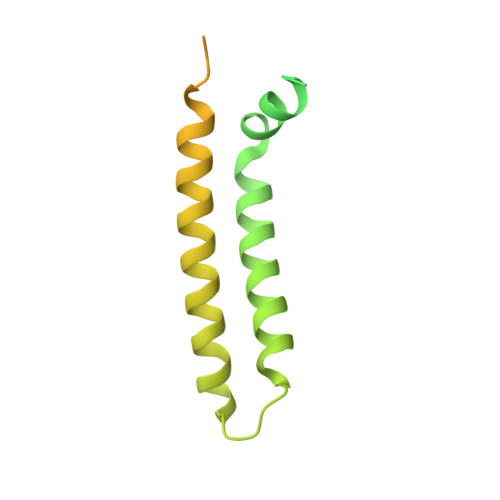
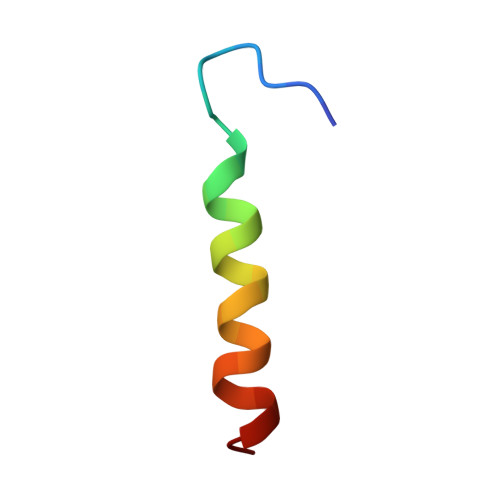
Cryo-EM structure and function of S. pombe complex IV with bound respiratory supercomplex factor.
Moe, A., Adelroth, P., Brzezinski, P., Nasvik Ojemyr, L.(2023) Commun Chem 6: 32-32
- PubMed: 36797353
- DOI: https://doi.org/10.1038/s42004-023-00827-3
- Primary Citation of Related Structures:
8C8Q - PubMed Abstract:
Fission yeast Schizosaccharomyces pombe serves as model organism for studying higher eukaryotes. We combined the use of cryo-EM and spectroscopy to investigate the structure and function of affinity purified respiratory complex IV (CIV) from S. pombe. The reaction sequence of the reduced enzyme with O 2 proceeds over a time scale of µs-ms, similar to that of the mammalian CIV. The cryo-EM structure of CIV revealed eleven subunits as well as a bound hypoxia-induced gene 1 (Hig1) domain of respiratory supercomplex factor 2 (Rcf2). These results suggest that binding of Rcf2 does not require the presence of a CIII-CIV supercomplex, i.e. Rcf2 is a component of CIV. An AlphaFold-Multimer model suggests that the Hig1 domains of both Rcf1 and Rcf2 bind at the same site of CIV suggesting that their binding is mutually exclusive. Furthermore, the differential functional effect of Rcf1 or Rcf2 is presumably caused by interactions of CIV with their different non-Hig1 domain parts.
- Department of Biochemistry and Biophysics, The Arrhenius Laboratories for Natural Sciences, Stockholm University, SE-106 91, Stockholm, Sweden.
Organizational Affiliation: