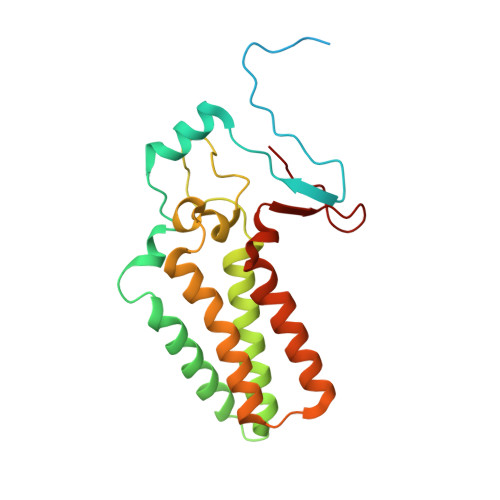
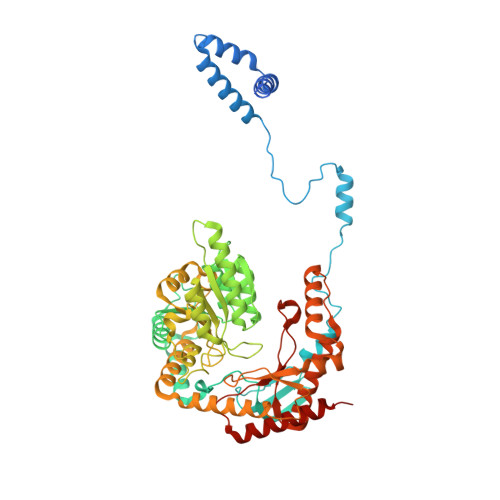
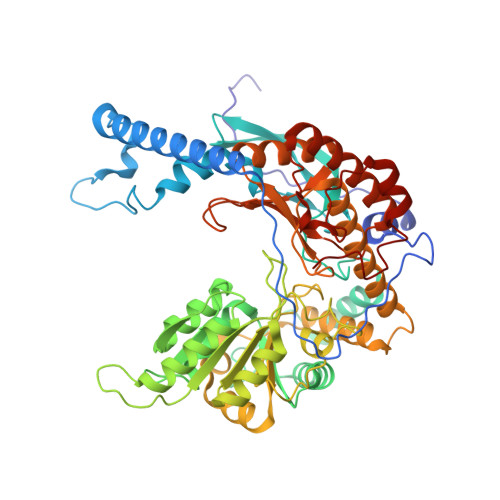
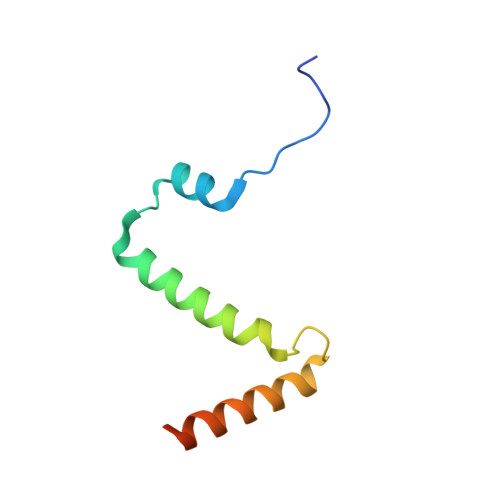
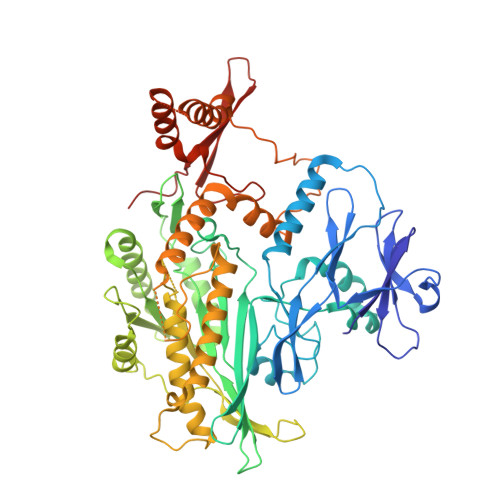
Structure of the ceramide-bound SPOTS complex.
Schafer, J.H., Korner, C., Esch, B.M., Limar, S., Parey, K., Walter, S., Januliene, D., Moeller, A., Frohlich, F.(2023) Nat Commun 14: 6196-6196
- PubMed: 37794019
- DOI: https://doi.org/10.1038/s41467-023-41747-z
- Primary Citation of Related Structures:
8C80, 8C81, 8C82 - PubMed Abstract:
Sphingolipids are structural membrane components that also function in cellular stress responses. The serine palmitoyltransferase (SPT) catalyzes the rate-limiting step in sphingolipid biogenesis. Its activity is tightly regulated through multiple binding partners, including Tsc3, Orm proteins, ceramides, and the phosphatidylinositol-4-phosphate (PI4P) phosphatase Sac1. The structural organization and regulatory mechanisms of this complex are not yet understood. Here, we report the high-resolution cryo-EM structures of the yeast SPT in complex with Tsc3 and Orm1 (SPOT) as dimers and monomers and a monomeric complex further carrying Sac1 (SPOTS). In all complexes, the tight interaction of the downstream metabolite ceramide and Orm1 reveals the ceramide-dependent inhibition. Additionally, observation of ceramide and ergosterol binding suggests a co-regulation of sphingolipid biogenesis and sterol metabolism within the SPOTS complex.
- Osnabrück University Department of Biology/Chemistry Structural Biology section, 49076, Osnabrück, Germany.
Organizational Affiliation: