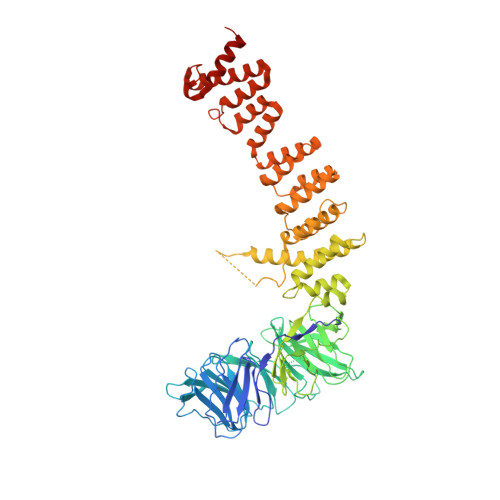
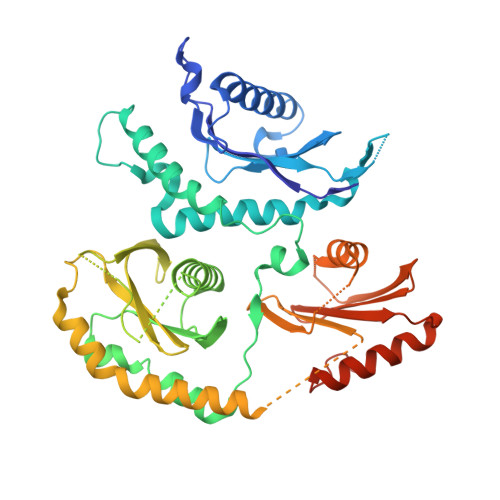
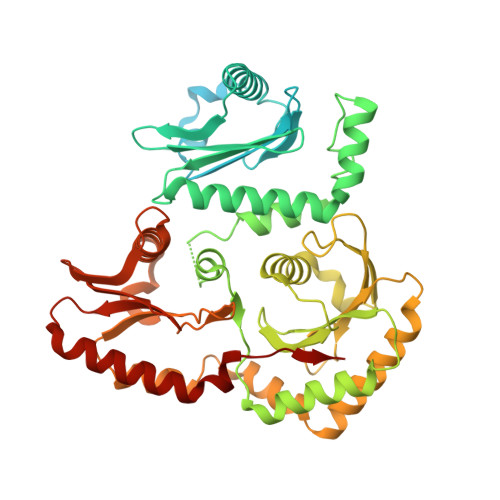
Structure of the metazoan Rab7 GEF complex Mon1-Ccz1-Bulli.
Herrmann, E., Schafer, J.H., Wilmes, S., Ungermann, C., Moeller, A., Kummel, D.(2023) Proc Natl Acad Sci U S A 120: e2301908120-e2301908120
- PubMed: 37155863
- DOI: https://doi.org/10.1073/pnas.2301908120
- Primary Citation of Related Structures:
8C7G - PubMed Abstract:
The endosomal system of eukaryotic cells represents a central sorting and recycling compartment linked to metabolic signaling and the regulation of cell growth. Tightly controlled activation of Rab GTPases is required to establish the different domains of endosomes and lysosomes. In metazoans, Rab7 controls endosomal maturation, autophagy, and lysosomal function. It is activated by the guanine nucleotide exchange factor (GEF) complex Mon1-Ccz1-Bulli (MCBulli) of the tri-longin domain (TLD) family. While the Mon1 and Ccz1 subunits have been shown to constitute the active site of the complex, the role of Bulli remains elusive. We here present the cryo-electron microscopy (cryo-EM) structure of MCBulli at 3.2 Å resolution. Bulli associates as a leg-like extension at the periphery of the Mon1 and Ccz1 heterodimers, consistent with earlier reports that Bulli does not impact the activity of the complex or the interactions with recruiter and substrate GTPases. While MCBulli shows structural homology to the related ciliogenesis and planar cell polarity effector (Fuzzy-Inturned-Wdpcp) complex, the interaction of the TLD core subunits Mon1-Ccz1 and Fuzzy-Inturned with Bulli and Wdpcp, respectively, is remarkably different. The variations in the overall architecture suggest divergent functions of the Bulli and Wdpcp subunits. Based on our structural analysis, Bulli likely serves as a recruitment platform for additional regulators of endolysosomal trafficking to sites of Rab7 activation.
- Department of Chemistry and Pharmacy, Institute of Biochemistry, University of Münster, 48149 Münster, Germany.
Organizational Affiliation: