Preclinical and Early Clinical Development of PTC596, a Novel Small-Molecule Tubulin-Binding Agent
Jernigan, F., Branstrom, A., Baird, J.D., Cao, L., Dali, M., Furia, B., Kim, M.J., O'Keefe, K., Kong, R., Laskin, O.L., Colacino, J.M., Pykett, M., Mollin, A., Sheedy, J., Dumble, M., Moon, Y.-C., Sheridan, R., Muehlethaler, T., Spiegel, R.J., Prota, A.E., Steinmetz, M.O., Weetall, M.(2021) Mol Cancer Ther 20: 1846-1857
- PubMed: 34315764
- DOI: https://doi.org/10.1158/1535-7163.MCT-20-0774
- Primary Citation of Related Structures:
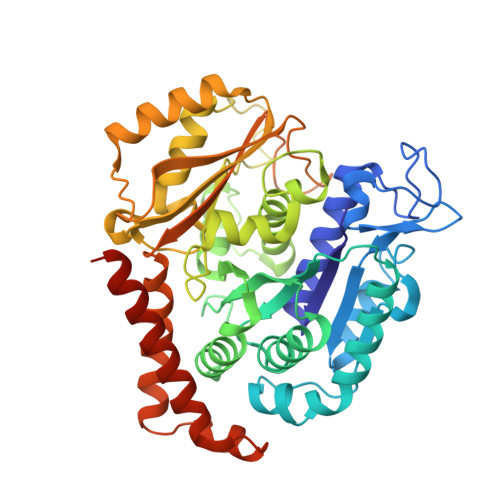
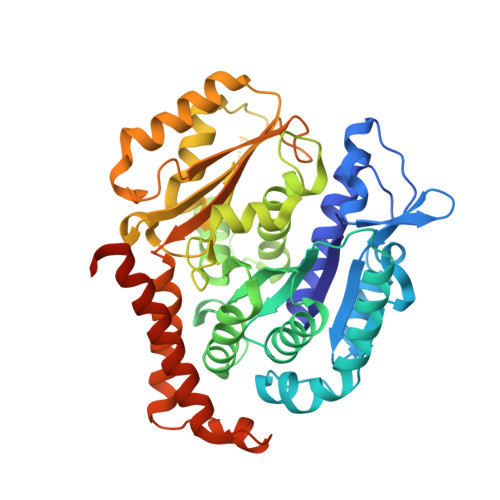
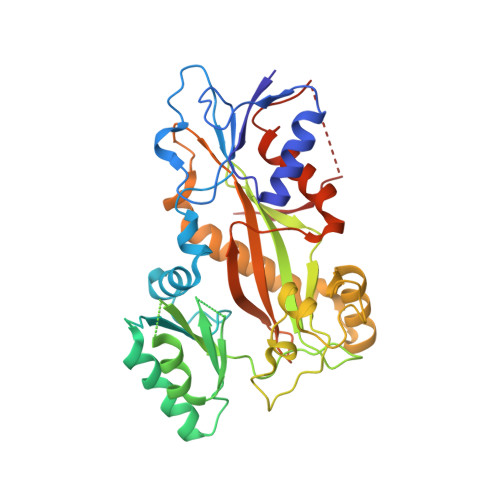
8C0F - PubMed Abstract:
PTC596 is an investigational small-molecule tubulin-binding agent. Unlike other tubulin-binding agents, PTC596 is orally bioavailable and is not a P-glycoprotein substrate. So as to characterize PTC596 to position the molecule for optimal clinical development, the interactions of PTC596 with tubulin using crystallography, its spectrum of preclinical in vitro anticancer activity, and its pharmacokinetic-pharmacodynamic relationship were investigated for efficacy in multiple preclinical mouse models of leiomyosarcomas and glioblastoma. Using X-ray crystallography, it was determined that PTC596 binds to the colchicine site of tubulin with unique key interactions. PTC596 exhibited broad-spectrum anticancer activity. PTC596 showed efficacy as monotherapy and additive or synergistic efficacy in combinations in mouse models of leiomyosarcomas and glioblastoma. PTC596 demonstrated efficacy in an orthotopic model of glioblastoma under conditions where temozolomide was inactive. In a first-in-human phase I clinical trial in patients with cancer, PTC596 monotherapy drug exposures were compared with those predicted to be efficacious based on mouse models. PTC596 is currently being tested in combination with dacarbazine in a clinical trial in adults with leiomyosarcoma and in combination with radiation in a clinical trial in children with diffuse intrinsic pontine glioma.
- PTC Therapeutics, Inc., South Plainfield, New Jersey.
Organizational Affiliation: