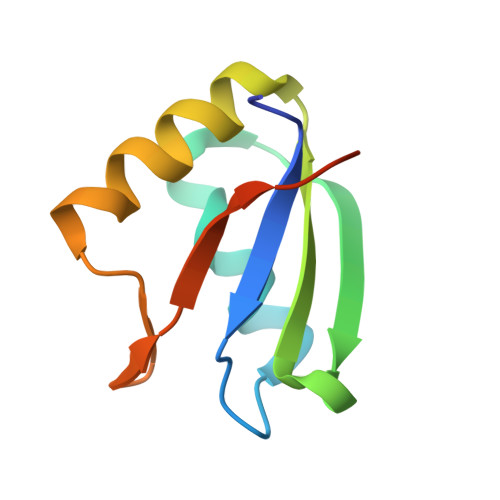
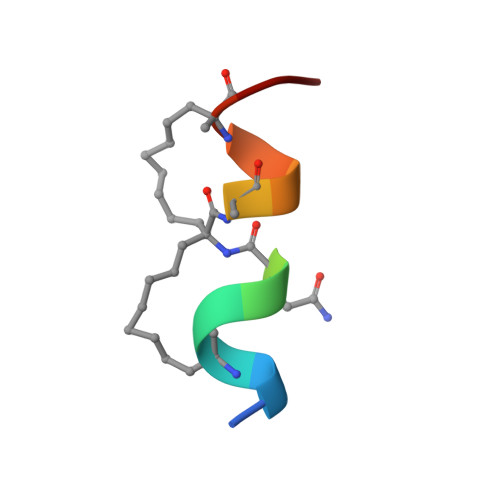
Rationally designed stapled peptides allosterically inhibit PTBP1-RNA-binding.
Schmeing, S., Amrahova, G., Bigler, K., Chang, J.Y., Openy, J., Pal, S., Posada, L., Gasper, R., 't Hart, P.(2023) Chem Sci 14: 8269-8278
- PubMed: 37564416
- DOI: https://doi.org/10.1039/d3sc00985h
- Primary Citation of Related Structures:
8BWF - PubMed Abstract:
The diverse role of the splicing factor PTBP1 in human cells has been widely studied and was found to be a driver for several diseases. PTBP1 binds RNA through its RNA-recognition motifs which lack obvious pockets for inhibition. A unique transient helix has been described to be part of its first RNA-recognition motif and to be important for RNA binding. In this study, we further confirmed the role of this helix and envisioned its dynamic nature as a unique opportunity to develop stapled peptide inhibitors of PTBP1. The peptides were found to be able to inhibit RNA binding via fluorescence polarization assays and directly occupy the helix binding site as observed by protein crystallography. These cell-permeable inhibitors were validated in cellulo to alter the regulation of alternative splicing events regulated by PTBP1. Our study demonstrates transient secondary structures of a protein can be mimicked by stapled peptides to inhibit allosteric mechanisms.
- Chemical Genomics Centre of the Max Planck Society, Max Planck Institute of Molecular Physiology Otto-Hahn-Strasse 11 44227 Dortmund Germany peter.t-hart@mpi-dortmund.mpg.de.
Organizational Affiliation: