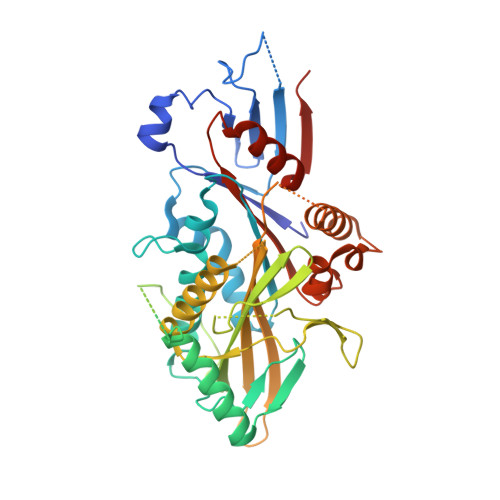
Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.
Ranaivoson, F.M., Crozet, V., Benoit, M.P.M.H., Abdalla Mohammed Khalid, A., Kikuti, C., Sirkia, H., El Marjou, A., Miserey-Lenkei, S., Asenjo, A.B., Sosa, H., Schmidt, C.F., Rosenfeld, S.S., Houdusse, A.(2023) Open Biol 13: 230122-230122
- PubMed: 37726093
- DOI: https://doi.org/10.1098/rsob.230122
- Primary Citation of Related Structures:
8BJS, 8F18, 8F1A - PubMed Abstract:
KIF20A is a critical kinesin for cell division and a promising anti-cancer drug target. The mechanisms underlying its cellular roles remain elusive. Interestingly, unusual coupling between the nucleotide- and microtubule-binding sites of this kinesin-6 has been reported, but little is known about how its divergent sequence leads to atypical motility properties. We present here the first high-resolution structure of its motor domain that delineates the highly unusual structural features of this motor, including a long L6 insertion that integrates into the core of the motor domain and that drastically affects allostery and ATPase activity. Together with the high-resolution cryo-electron microscopy microtubule-bound KIF20A structure that reveals the microtubule-binding interface, we dissect the peculiarities of the KIF20A sequence that influence its mechanochemistry, leading to low motility compared to other kinesins. Structural and functional insights from the KIF20A pre-power stroke conformation highlight the role of extended insertions in shaping the motor's mechanochemical cycle. Essential for force production and processivity is the length of the neck linker in kinesins. We highlight here the role of the sequence preceding the neck linker in controlling its backward docking and show that a neck linker four times longer than that in kinesin-1 is required for the activity of this motor.
- Structural Motility, CNRS UMR144, Institut Curie, Université Paris Sciences et Lettres, Sorbonne Université, 75248 Paris, France.
Organizational Affiliation: