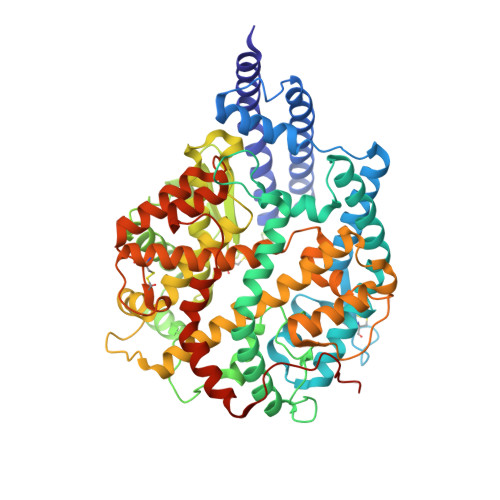
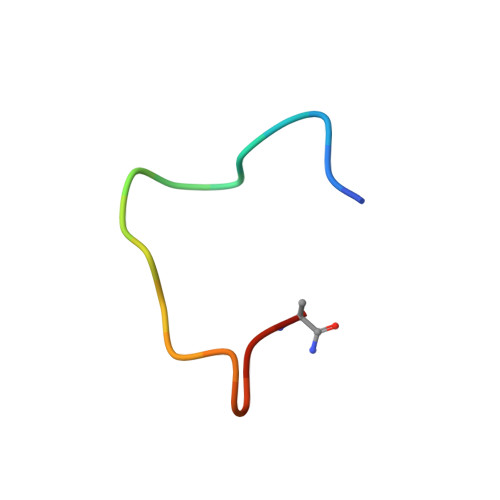
Structure-Guided Chemical Optimization of Bicyclic Peptide ( Bicycle ) Inhibitors of Angiotensin-Converting Enzyme 2.
Harman, M.A.J., Stanway, S.J., Scott, H., Demydchuk, Y., Bezerra, G.A., Pellegrino, S., Chen, L., Brear, P., Lulla, A., Hyvonen, M., Beswick, P.J., Skynner, M.J.(2023) J Med Chem 66: 9881-9893
- PubMed: 37433017
- DOI: https://doi.org/10.1021/acs.jmedchem.3c00710
- Primary Citation of Related Structures:
8B9P, 8BFW, 8BN1, 8BYJ - PubMed Abstract:
Angiotensin-converting enzyme 2 (ACE2) is a metalloprotease that cleaves angiotensin II, a peptide substrate involved in the regulation of hypertension. Here, we identified a series of constrained bicyclic peptides, Bicycle , inhibitors of human ACE2 by panning highly diverse bacteriophage display libraries. These were used to generate X-ray crystal structures which were used to inform the design of additional Bicycles with increased affinity and inhibition of ACE2 enzymatic activity. This novel structural class of ACE2 inhibitors is among the most potent ACE2 inhibitors yet described in vitro , representing a valuable tool to further probe ACE2 function and for potential therapeutic utility.
- BicycleTx Ltd., Portway Building Blocks A and B, Granta Park, Great Abington, Cambridge CB21 6GS, U.K.
Organizational Affiliation: