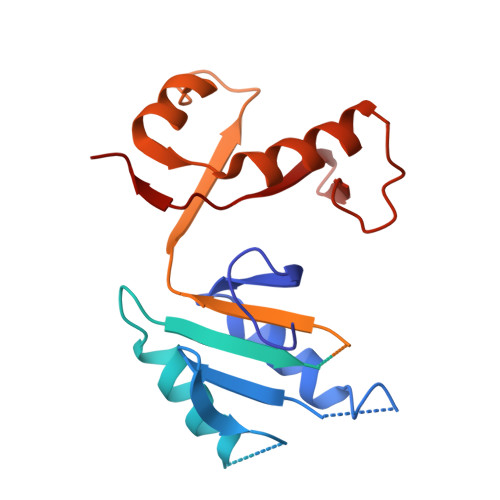
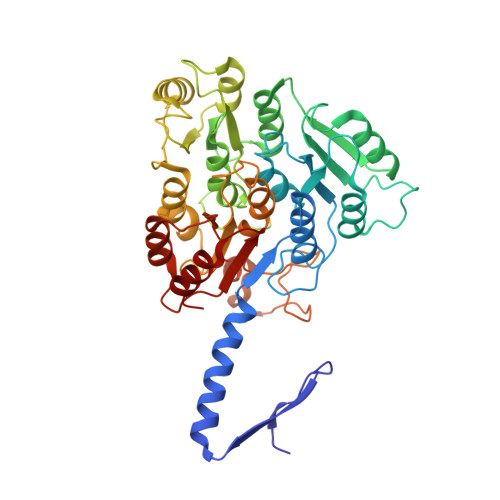
An intact S-layer is advantageous to Clostridioides difficile within the host.
Ormsby, M.J., Vaz, F., Kirk, J.A., Barwinska-Sendra, A., Hallam, J.C., Lanzoni-Mangutchi, P., Cole, J., Chaudhuri, R.R., Salgado, P.S., Fagan, R.P., Douce, G.R.(2023) PLoS Pathog 19: e1011015-e1011015
- PubMed: 37384772
- DOI: https://doi.org/10.1371/journal.ppat.1011015
- Primary Citation of Related Structures:
8BBY - PubMed Abstract:
Clostridioides difficile is responsible for substantial morbidity and mortality in antibiotically-treated, hospitalised, elderly patients, in which toxin production correlates with diarrhoeal disease. While the function of these toxins has been studied in detail, the contribution of other factors, including the paracrystalline surface layer (S-layer), to disease is less well understood. Here, we highlight the essentiality of the S-layer in vivo by reporting the recovery of S-layer variants, following infection with the S-layer-null strain, FM2.5. These variants carry either correction of the original point mutation, or sequence modifications which restored the reading frame, and translation of slpA. Selection of these variant clones was rapid in vivo, and independent of toxin production, with up to 90% of the recovered C. difficile population encoding modified slpA sequence within 24 h post infection. Two variants, subsequently named FM2.5varA and FM2.5varB, were selected for study in greater detail. Structural determination of SlpA from FM2.5varB indicated an alteration in the orientation of protein domains, resulting in a reorganisation of the lattice assembly, and changes in interacting interfaces, which might alter function. Interestingly, variant FM2.5varB displayed an attenuated, FM2.5-like phenotype in vivo compared to FM2.5varA, which caused disease severity more comparable to that of R20291. Comparative RNA sequencing (RNA-Seq) analysis of in vitro grown isolates revealed large changes in gene expression between R20291 and FM2.5. Downregulation of tcdA/tcdB and several genes associated with sporulation and cell wall integrity may account for the reported attenuated phenotype of FM2.5 in vivo. RNA-seq data correlated well with disease severity with the more virulent variant, FM2.5varA, showing s similar profile of gene expression to R20291 in vitro, while the attenuated FM2.5varB showed downregulation of many of the same virulence associated traits as FM2.5. Cumulatively, these data add to a growing body of evidence that the S-layer contributes to C. difficile pathogenesis and disease severity.
- School of Infection and Immunity, College of Medical, Veterinary & Life Sciences, University of Glasgow, Scotland, United Kingdom.
Organizational Affiliation: