Switching of Receptor Binding Poses between Closely Related Enteroviruses.
Zhou, D., Qin, L., Duyvesteyn, H.M.E., Zhao, Y., Lin, T.Y., Fry, E.E., Ren, J., Huang, K.A., Stuart, D.I.(2022) Viruses 14
- PubMed: 36560629
- DOI: https://doi.org/10.3390/v14122625
- Primary Citation of Related Structures:
8B8R, 8B9F - PubMed Abstract:
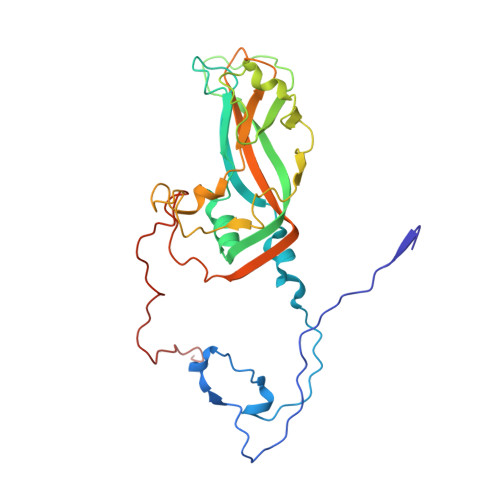
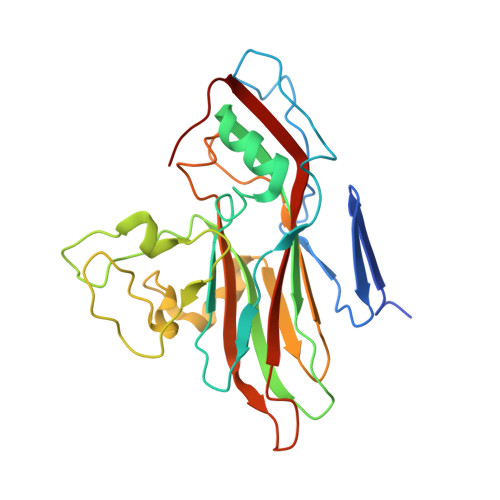
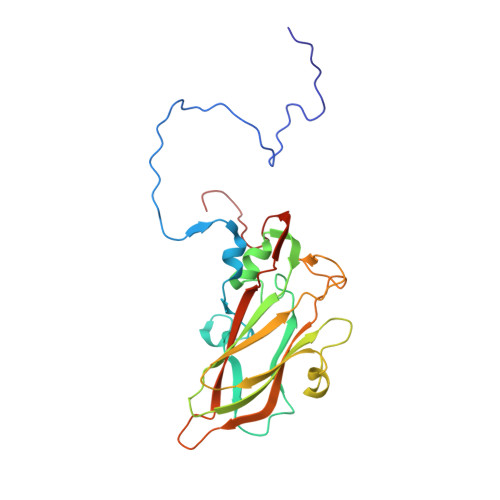
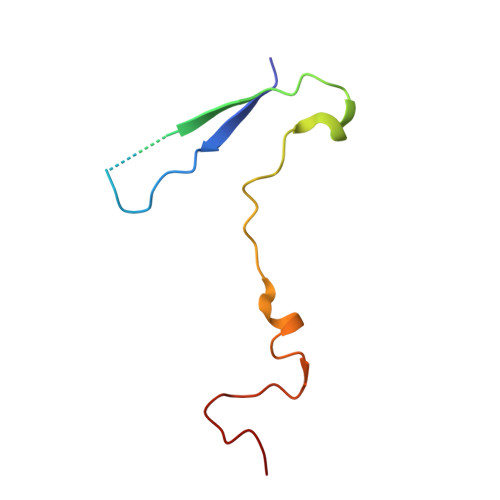
Echoviruses, for which there are currently no approved vaccines or drugs, are responsible for a range of human diseases, for example echovirus 11 (E11) is a major cause of serious neonatal morbidity and mortality. Decay-accelerating factor (DAF, also known as CD55) is an attachment receptor for E11. Here, we report the structure of the complex of E11 and the full-length ectodomain of DAF (short consensus repeats, SCRs, 1-4) at 3.1 Å determined by cryo-electron microscopy (cryo-EM). SCRs 3 and 4 of DAF interact with E11 at the southern rim of the canyon via the VP2 EF and VP3 BC loops. We also observe an unexpected interaction between the N-linked glycan (residue 95 of DAF) and the VP2 BC loop of E11. DAF is a receptor for at least 20 enteroviruses and we classify its binding patterns from reported DAF/virus complexes into two distinct positions and orientations, named as E6 and E11 poses. Whilst 60 DAF molecules can attach to the virion in the E6 pose, no more than 30 can attach to E11 due to steric restrictions. Analysis of the distinct modes of interaction and structure and sequence-based phylogenies suggests that the two modes evolved independently, with the E6 mode likely found earlier.
- Division of Structural Biology, Nuffield Department of Medicine, University of Oxford, The Wellcome Centre for Human Genetics, Headington, Oxford OX3 7BN, UK.
Organizational Affiliation: