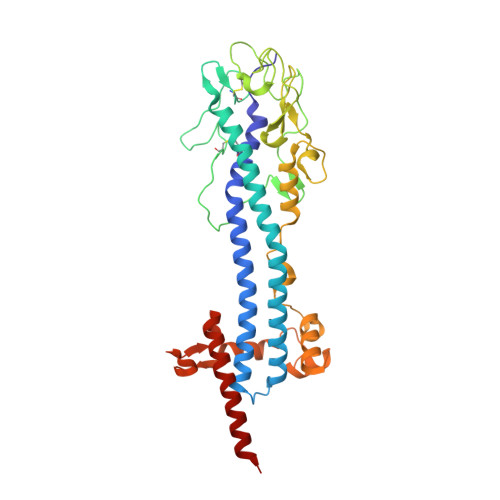
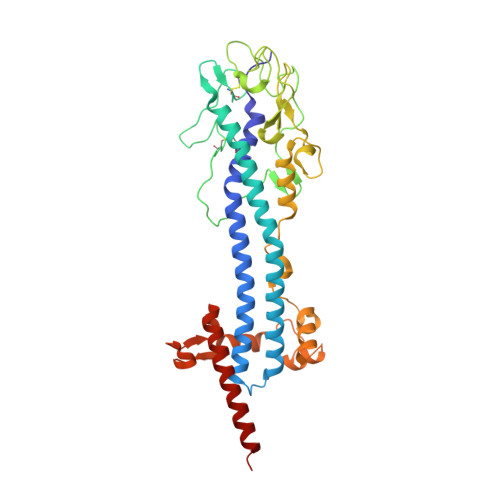
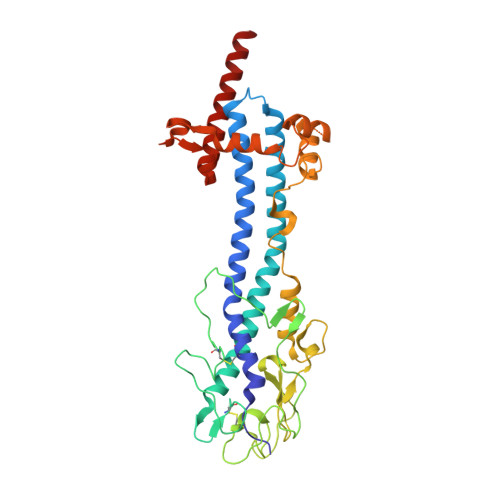
Structural similarities between the metacyclic and bloodstream form variant surface glycoproteins of the African trypanosome.
Chandra, M., Dakovic, S., Foti, K., Zeelen, J.P., van Straaten, M., Aresta-Branco, F., Tihon, E., Lubbehusen, N., Ruppert, T., Glover, L., Papavasiliou, F.N., Stebbins, C.E.(2023) PLoS Negl Trop Dis 17: e0011093-e0011093
- PubMed: 36780870
- DOI: https://doi.org/10.1371/journal.pntd.0011093
- Primary Citation of Related Structures:
8B3B, 8B3E, 8B3W - PubMed Abstract:
During infection of mammalian hosts, African trypanosomes thwart immunity using antigenic variation of the dense Variant Surface Glycoprotein (VSG) coat, accessing a large repertoire of several thousand genes and pseudogenes, and switching to antigenically distinct copies. The parasite is transferred to mammalian hosts by the tsetse fly. In the salivary glands of the fly, the pathogen adopts the metacyclic form and expresses a limited repertoire of VSG genes specific to that developmental stage. It has remained unknown whether the metacyclic VSGs possess distinct properties associated with this particular and discrete phase of the parasite life cycle. We present here three novel metacyclic form VSG N-terminal domain crystal structures (mVSG397, mVSG531, and mVSG1954) and show that they mirror closely in architecture, oligomerization, and surface diversity the known classes of bloodstream form VSGs. These data suggest that the mVSGs are unlikely to be a specialized subclass of VSG proteins, and thus could be poor candidates as the major components of prophylactic vaccines against trypanosomiasis.
- Division of Structural Biology of Infection and Immunity, German Cancer Research Center, Heidelberg, Germany.
Organizational Affiliation: