Crystal structure determination of a highly active UDP-glucose pyrophosphorylase from Thermocrispum agreste DSM 44070
Kumpf, A., Maier, A., Laustsen, J.U., Jeffries, C.M., Bento, I., Tischler, D.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UTP--glucose-1-phosphate uridylyltransferase | 299 | Thermocrispum agreste DSM 44070 | Mutation(s): 0 Gene Names: DIU77_00305 EC: 2.7.7.9 | 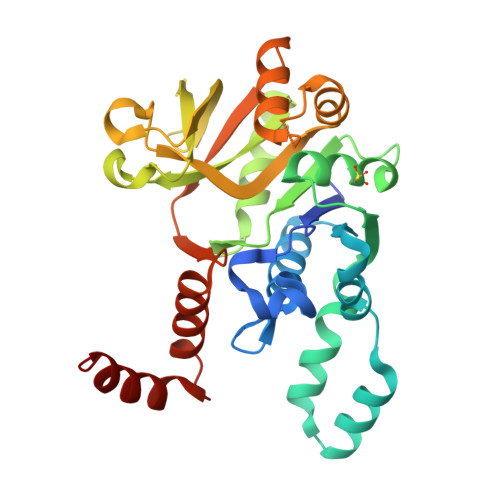 | |
UniProt | |||||
Find proteins for A0A2W4LV58 (Thermocrispum agreste) Explore A0A2W4LV58 Go to UniProtKB: A0A2W4LV58 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2W4LV58 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | C [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | D [auth A] E [auth A] F [auth A] G [auth A] H [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| SCN Query on SCN | N [auth A] | THIOCYANATE ION C N S ZMZDMBWJUHKJPS-UHFFFAOYSA-M |  | ||
| K Query on K | O [auth A], P [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSX Query on CSX | A, B | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.086 | α = 90 |
| b = 78.593 | β = 93.885 |
| c = 89.788 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| European Union (EU) | European Union | 100263899 |