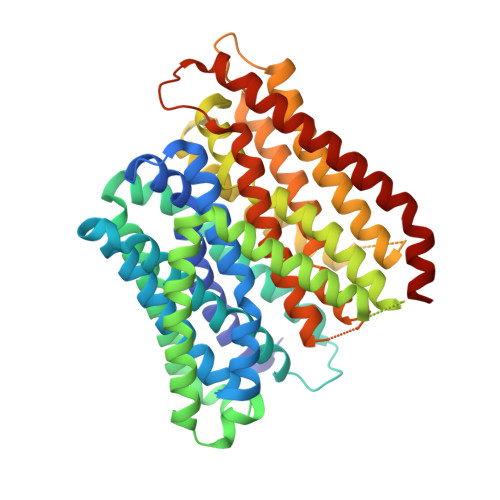
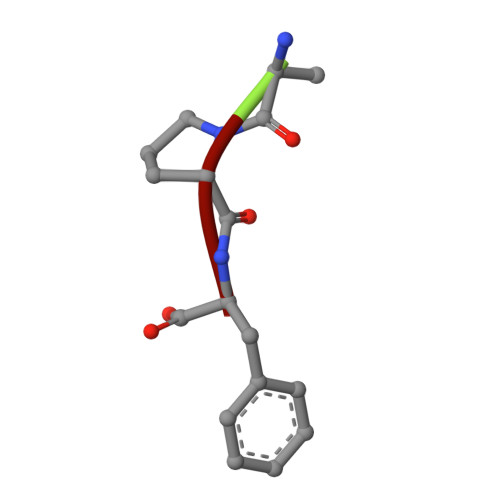
Plasticity of the binding pocket in peptide transporters underpins promiscuous substrate recognition.
Kotov, V., Killer, M., Jungnickel, K.E.J., Lei, J., Finocchio, G., Steinke, J., Bartels, K., Strauss, J., Dupeux, F., Humm, A.S., Cornaciu, I., Marquez, J.A., Pardon, E., Steyaert, J., Low, C.(2023) Cell Rep 42: 112831-112831
- PubMed: 37467108
- DOI: https://doi.org/10.1016/j.celrep.2023.112831
- Primary Citation of Related Structures:
8B17, 8B18, 8B19, 8B1A, 8B1B, 8B1C, 8B1D, 8B1E, 8B1F, 8B1G, 8B1H, 8B1I, 8B1J, 8B1K - PubMed Abstract:
Proton-dependent oligopeptide transporters (POTs) are promiscuous transporters of the major facilitator superfamily that constitute the main route of entry for a wide range of dietary peptides and orally administrated peptidomimetic drugs. Given their clinical and pathophysiological relevance, several POT homologs have been studied extensively at the structural and molecular level. However, the molecular basis of recognition and transport of diverse peptide substrates has remained elusive. We present 14 X-ray structures of the bacterial POT DtpB in complex with chemically diverse di- and tripeptides, providing novel insights into the plasticity of the conserved central binding cavity. We analyzed binding affinities for more than 80 peptides and monitored uptake by a fluorescence-based transport assay. To probe whether all 8400 natural di- and tripeptides can bind to DtpB, we employed state-of-the-art molecular docking and machine learning and conclude that peptides with compact hydrophobic residues are the best DtpB binders.
- Center for Structural Systems Biology (CSSB), Notkestraße 85, 22607 Hamburg, Germany; European Molecular Biology Laboratory (EMBL) Hamburg, Notkestraße 85, 22607 Hamburg, Germany.
Organizational Affiliation: