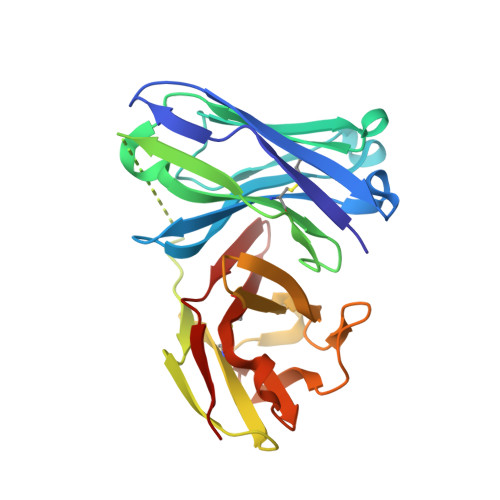
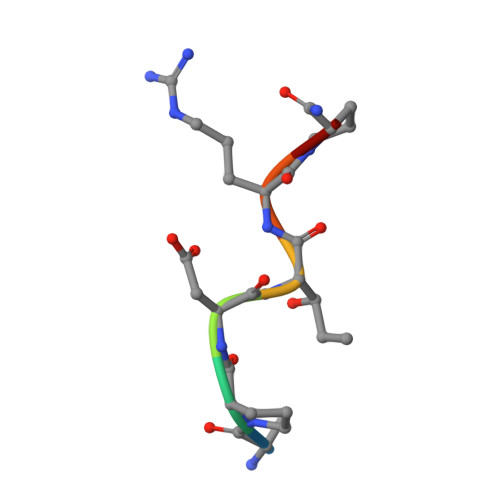
Structure-Guided Approach for the Development of MUC1-Glycopeptide-Based Cancer Vaccines with Predictable Responses.
Bermejo, I.A., Guerreiro, A., Eguskiza, A., Martinez-Saez, N., Lazaris, F.S., Asin, A., Somovilla, V.J., Companon, I., Raju, T.K., Tadic, S., Garrido, P., Garcia-Sanmartin, J., Mangini, V., Grosso, A.S., Marcelo, F., Avenoza, A., Busto, J.H., Garcia-Martin, F., Hurtado-Guerrero, R., Peregrina, J.M., Bernardes, G.J.L., Martinez, A., Fiammengo, R., Corzana, F.(2024) JACS Au 4: 150-163
- PubMed: 38274250
- DOI: https://doi.org/10.1021/jacsau.3c00587
- Primary Citation of Related Structures:
8AXH - PubMed Abstract:
Mucin-1 (MUC1) glycopeptides are exceptional candidates for potential cancer vaccines. However, their autoantigenic nature often results in a weak immune response. To overcome this drawback, we carefully engineered synthetic antigens with precise chemical modifications. To be effective and stimulate an anti-MUC1 response, artificial antigens must mimic the conformational dynamics of natural antigens in solution and have an equivalent or higher binding affinity to anti-MUC1 antibodies than their natural counterparts. As a proof of concept, we have developed a glycopeptide that contains noncanonical amino acid (2 S ,3 R )-3-hydroxynorvaline. The unnatural antigen fulfills these two properties and effectively mimics the threonine-derived antigen. On the one hand, conformational analysis in water shows that this surrogate explores a landscape similar to that of the natural variant. On the other hand, the presence of an additional methylene group in the side chain of this analog compared to the threonine residue enhances a CH/π interaction in the antigen/antibody complex. Despite an enthalpy-entropy balance, this synthetic glycopeptide has a binding affinity slightly higher than that of its natural counterpart. When conjugated with gold nanoparticles, the vaccine candidate stimulates the formation of specific anti-MUC1 IgG antibodies in mice and shows efficacy comparable to that of the natural derivative. The antibodies also exhibit cross-reactivity to selectively target, for example, human breast cancer cells. This investigation relied on numerous analytical (e.g., NMR spectroscopy and X-ray crystallography) and biophysical techniques and molecular dynamics simulations to characterize the antigen-antibody interactions. This workflow streamlines the synthetic process, saves time, and reduces the need for extensive, animal-intensive immunization procedures. These advances underscore the promise of structure-based rational design in the advance of cancer vaccine development.
- Department of Chemistry and Instituto de Investigación en Química de la Universidad de La Rioja (IQUR), Universidad de La Rioja, Logroño 26006, Spain.
Organizational Affiliation: