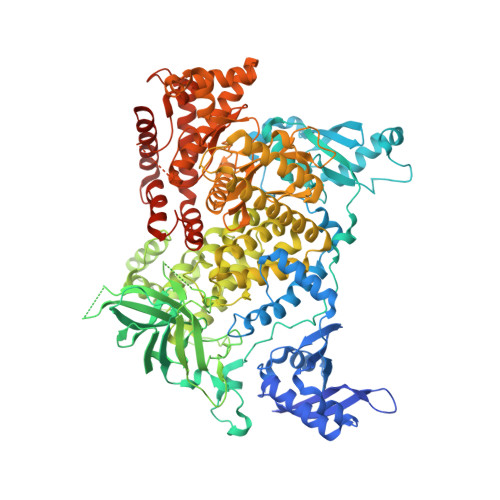
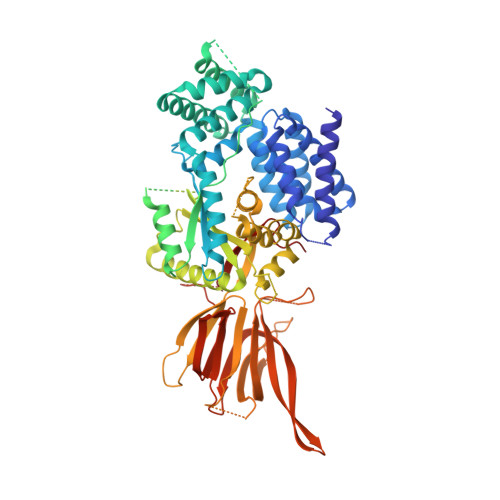
Molecular basis for differential activation of p101 and p84 complexes of PI3K gamma by Ras and GPCRs.
Rathinaswamy, M.K., Jenkins, M.L., Duewell, B.R., Zhang, X., Harris, N.J., Evans, J.T., Stariha, J.T.B., Dalwadi, U., Fleming, K.D., Ranga-Prasad, H., Yip, C.K., Williams, R.L., Hansen, S.D., Burke, J.E.(2023) Cell Rep 42: 112172-112172
- PubMed: 36842083
- DOI: https://doi.org/10.1016/j.celrep.2023.112172
- Primary Citation of Related Structures:
8AJ8 - PubMed Abstract:
Class IB phosphoinositide 3-kinase (PI3Kγ) is activated in immune cells and can form two distinct complexes (p110γ-p84 and p110γ-p101), which are differentially activated by G protein-coupled receptors (GPCRs) and Ras. Using a combination of X-ray crystallography, hydrogen deuterium exchange mass spectrometry (HDX-MS), electron microscopy, molecular modeling, single-molecule imaging, and activity assays, we identify molecular differences between p110γ-p84 and p110γ-p101 that explain their differential membrane recruitment and activation by Ras and GPCRs. The p110γ-p84 complex is dynamic compared with p110γ-p101. While p110γ-p101 is robustly recruited by Gβγ subunits, p110γ-p84 is weakly recruited to membranes by Gβγ subunits alone and requires recruitment by Ras to allow for Gβγ activation. We mapped two distinct Gβγ interfaces on p101 and the p110γ helical domain, with differences in the C-terminal domain of p84 and p101 conferring sensitivity of p110γ-p101 to Gβγ activation. Overall, our work provides key insight into the molecular basis for how PI3Kγ complexes are activated.
- Department of Biochemistry and Microbiology, University of Victoria, Victoria, BC V8W 2Y2, Canada.
Organizational Affiliation: