A Systematic Approach to the Discovery of Protein-Protein Interaction Stabilizers.
Kenanova, D.N., Visser, E.J., Virta, J.M., Sijbesma, E., Centorrino, F., Vickery, H.R., Zhong, M., Neitz, R.J., Brunsveld, L., Ottmann, C., Arkin, M.R.(2023) ACS Cent Sci 9: 937-946
- PubMed: 37252362
- DOI: https://doi.org/10.1021/acscentsci.2c01449
- Primary Citation of Related Structures:
8A62, 8A65, 8A68, 8A6F, 8A6H, 8ADM, 8AFN, 8AV0 - PubMed Abstract:
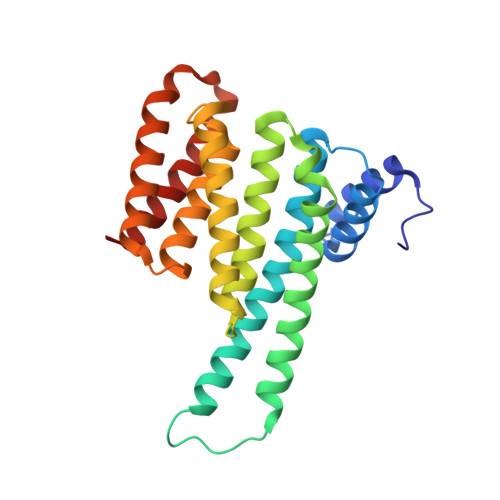
Dysregulation of protein-protein interactions (PPIs) commonly leads to disease. PPI stabilization has only recently been systematically explored for drug discovery despite being a powerful approach to selectively target intrinsically disordered proteins and hub proteins, like 14-3-3, with multiple interaction partners. Disulfide tethering is a site-directed fragment-based drug discovery (FBDD) methodology for identifying reversibly covalent small molecules. We explored the scope of disulfide tethering for the discovery of selective PPI stabilizers (molecular glues) using the hub protein 14-3-3σ. We screened complexes of 14-3-3 with 5 biologically and structurally diverse phosphopeptides derived from the 14-3-3 client proteins ERα, FOXO1, C-RAF, USP8, and SOS1. Stabilizing fragments were found for 4/5 client complexes. Structural elucidation of these complexes revealed the ability of some peptides to conformationally adapt to make productive interactions with the tethered fragments. We validated eight fragment stabilizers, six of which showed selectivity for one phosphopeptide client, and structurally characterized two nonselective hits and four fragments that selectively stabilized C-RAF or FOXO1. The most efficacious fragment increased 14-3-3σ/C-RAF phosphopeptide affinity by 430-fold. Disulfide tethering to the wildtype C38 in 14-3-3σ provided diverse structures for future optimization of 14-3-3/client stabilizers and highlighted a systematic method to discover molecular glues.
- Department of Pharmaceutical Chemistry and Small Molecule Discovery Center (SMDC), University of California, San Francisco 94143, United States.
Organizational Affiliation: