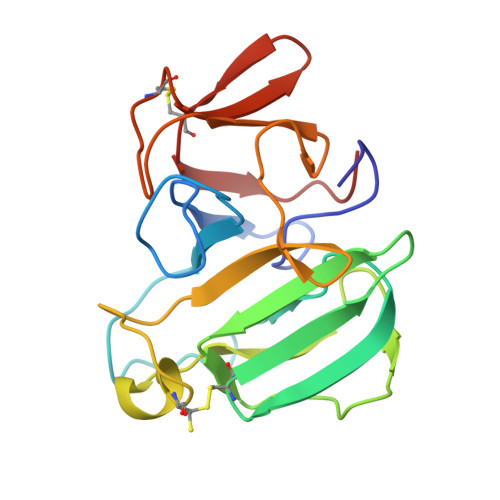
Structural and Functional Characterization of beta-lytic Protease from Lysobacter capsici VKM B-2533 T.
Afoshin, A., Tishchenko, S., Gabdulkhakov, A., Kudryakova, I., Galemina, I., Zelenov, D., Leontyevskaya, E., Saharova, S., Leontyevskaya Vasilyeva, N.(2022) Int J Mol Sci 23
- PubMed: 36555752
- DOI: https://doi.org/10.3390/ijms232416100
- Primary Citation of Related Structures:
8AF1 - PubMed Abstract:
The crystal structure of the Lysobacter capsici VKM B-2533 T β-lytic protease (Blp), a medicinally promising antimicrobial enzyme, was first solved. Blp was established to possess a folding characteristic of the M23 protease family. The groove of the Blp active site, as compared with that of the LasA structural homologue from Pseudomonas aeruginosa , was found to have amino acid differences. Biochemical analysis revealed no differences in the optimal reaction conditions for manifesting Blp and LasA bacteriolytic activities. At the same time, Blp had a broader range of action against living and autoclaved target cells. The results suggest that the distinction in the geometry of the active site and the charge of amino acid residues that form the active site groove can be important for the hydrolysis of different peptidoglycan types in target cells.
- Laboratory of Microbial Cell Surface Biochemistry, G.K. Skryabin Institute of Biochemistry and Physiology of Microorganisms, FRC PSCBR, Russian Academy of Sciences, 5 Prosp. Nauki, 142290 Pushchino, Russia.
Organizational Affiliation: