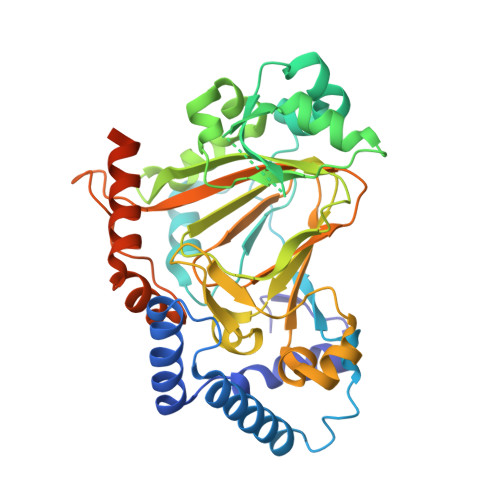
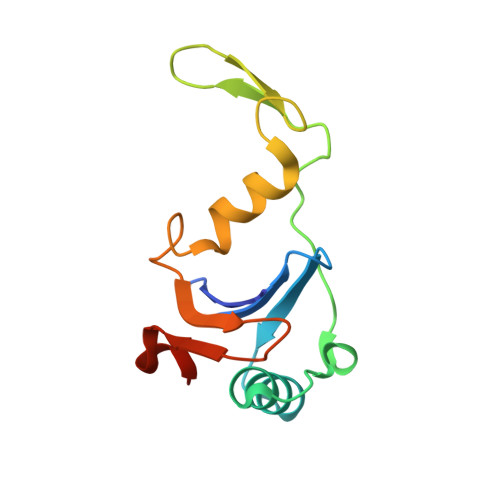
Heterocomplex structure of a polyketide synthase component involved in modular backbone halogenation.
Fraley, A.E., Dell, M., Schmalhofer, M., Meoded, R.A., Bergande, C., Groll, M., Piel, J.(2023) Structure 31: 565
- PubMed: 36917986
- DOI: https://doi.org/10.1016/j.str.2023.02.010
- Primary Citation of Related Structures:
8A82 - PubMed Abstract:
Bacterial modular polyketide synthases (PKSs) generate diverse, complex and bioactive natural products that are constructed mainly based on principles of fatty acid biosynthesis. The cytotoxic oocydin-type polyketides contain a vinyl chloride moiety introduced during polyketide chain elongation. Required for modular polyketide backbone halogenation are a non-heme iron and ɑ-ketoglutarate-dependent halogenase OocP and OocQ lacking characterized homologs. This work provides structural insights into these unusual PKS components and their interactions via a high-resolution X-ray crystallography structure of the heterocomplex. By mapping the protein-protein interactions and comparison with structures of similar halogenases, we illustrate the potential of this heterodimer complex as a replacement for the conserved homodimeric structure of homologous enzymes. The OocPQ protein pair has thus evolved as a means of stabilizing the halogenase and facilitating chemical transformations with great synthetic utility.
- Institute of Microbiology, Eidgenössische Technische Hochschule (ETH) Zurich, Zurich, Switzerland.
Organizational Affiliation: