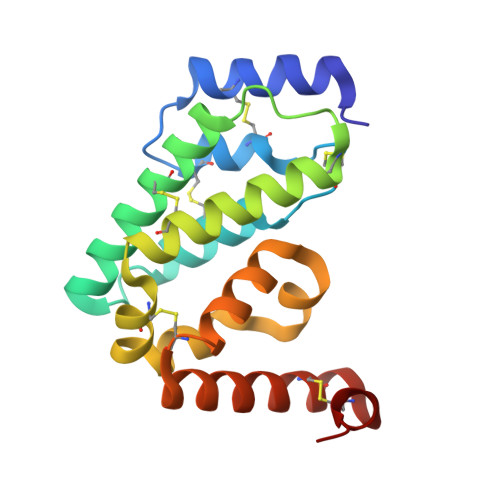
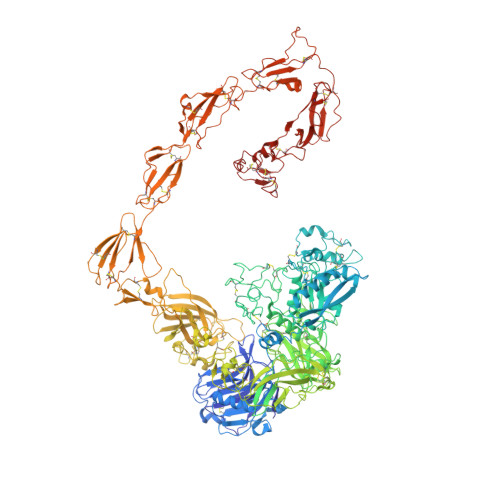
Structure of the proteolytic enzyme PAPP-A with the endogenous inhibitor stanniocalcin-2 reveals its inhibitory mechanism.
Kobbero, S.D., Gajhede, M., Mirza, O.A., Kloverpris, S., Kjaer, T.R., Mikkelsen, J.H., Boesen, T., Oxvig, C.(2022) Nat Commun 13: 6084-6084
- PubMed: 36257932
- DOI: https://doi.org/10.1038/s41467-022-33698-8
- Primary Citation of Related Structures:
8A7D, 8A7E - PubMed Abstract:
The metzincin metalloproteinase PAPP-A plays a key role in the regulation of insulin-like growth factor (IGF) signaling by specific cleavage of inhibitory IGF binding proteins (IGFBPs). Using single-particle cryo-electron microscopy (cryo-EM), we here report the structure of PAPP-A in complex with its endogenous inhibitor, stanniocalcin-2 (STC2), neither of which have been reported before. The highest resolution (3.1 Å) was obtained for the STC2 subunit and the N-terminal approximately 1000 residues of the PAPP-A subunit. The 500 kDa 2:2 PAPP-A·STC2 complex is a flexible multidomain ensemble with numerous interdomain contacts. In particular, a specific disulfide bond between the subunits of STC2 and PAPP-A prevents dissociation, and interactions between STC2 and a module located in the very C-terminal end of the PAPP-A subunit prevent binding of its main substrate, IGFBP-4. While devoid of activity towards IGFBP-4, the active site cleft of the catalytic domain is accessible in the inhibited PAPP-A·STC2 complex, as shown by its ability to hydrolyze a synthetic peptide derived from IGFBP-4. Relevant to multiple human pathologies, this unusual mechanism of proteolytic inhibition may support the development of specific pharmaceutical agents, by which IGF signaling can be indirectly modulated.
- Department of Molecular Biology and Genetics, Aarhus University, DK-8000, Aarhus C, Denmark.
Organizational Affiliation: