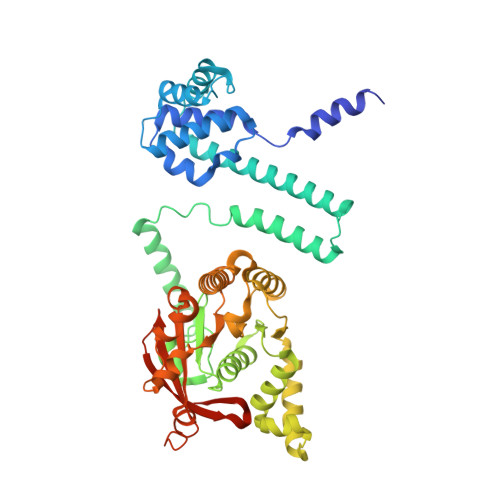
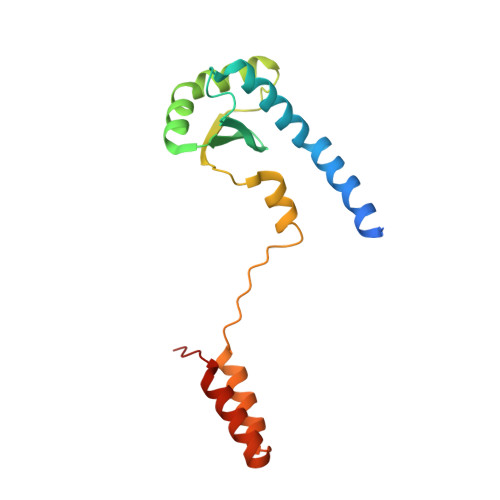
The LH-DH module of bacterial replicative helicases is the common binding site for DciA and other helicase loaders.
Cargemel, C., Marsin, S., Noiray, M., Legrand, P., Bounoua, H., Li de la Sierra-Gallay, I., Walbott, H., Quevillon-Cheruel, S.(2023) Acta Crystallogr D Struct Biol 79: 177-187
- PubMed: 36762863
- DOI: https://doi.org/10.1107/S2059798323000281
- Primary Citation of Related Structures:
8A3V - PubMed Abstract:
During the initiation step of bacterial genome replication, replicative helicases depend on specialized proteins for their loading onto oriC. DnaC and DnaI were the first loaders to be characterized. However, most bacteria do not contain any of these genes, which are domesticated phage elements that have replaced the ancestral and unrelated loader gene dciA several times during evolution. To understand how DciA assists the loading of DnaB, the crystal structure of the complex from Vibrio cholerae was determined, in which two VcDciA molecules interact with a dimer of VcDnaB without changing its canonical structure. The data showed that the VcDciA binding site on VcDnaB is the conserved module formed by the linker helix LH of one monomer and the determinant helix DH of the second monomer. Interestingly, DnaC from Escherichia coli also targets this module onto EcDnaB. Thanks to their common target site, it was shown that VcDciA and EcDnaC could be functionally interchanged in vitro despite sharing no structural similarity. This represents a milestone in understanding the mechanism employed by phage helicase loaders to hijack bacterial replicative helicases during evolution.
- Université Paris-Saclay, CEA, CNRS, Institute for Integrative Biology of the Cell (I2BC), 91180 Gif-sur-Yvette, France.
Organizational Affiliation: