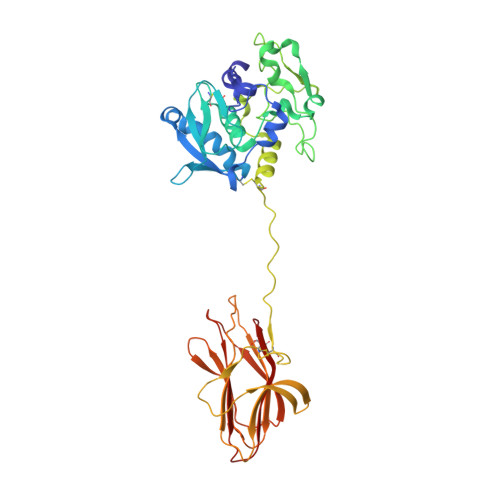
Zymogenic latency in an ∼250-million-year-old astacin metallopeptidase.
Guevara, T., Rodriguez-Banqueri, A., Stocker, W., Becker-Pauly, C., Gomis-Ruth, F.X.(2022) Acta Crystallogr D Struct Biol 78: 1347-1357
- PubMed: 36322418
- DOI: https://doi.org/10.1107/S2059798322009688
- Primary Citation of Related Structures:
8A28 - PubMed Abstract:
The horseshoe crab Limulus polyphemus is one of few extant Limulus species, which date back to ∼250 million years ago under the conservation of a common Bauplan documented by fossil records. It possesses the only proteolytic blood-coagulation and innate immunity system outside vertebrates and is a model organism for the study of the evolution and function of peptidases. The astacins are a family of metallopeptidases that share a central ∼200-residue catalytic domain (CD), which is found in >1000 species across holozoans and, sporadically, bacteria. Here, the zymogen of an astacin from L. polyphemus was crystallized and its structure was solved. A 34-residue, mostly unstructured pro-peptide (PP) traverses, and thus blocks, the active-site cleft of the CD in the opposite direction to a substrate. A central `PP motif' (F 35 -E-G-D-I 39 ) adopts a loop structure which positions Asp38 to bind the catalytic metal, replacing the solvent molecule required for catalysis in the mature enzyme according to an `aspartate-switch' mechanism. Maturation cleavage of the PP liberates the cleft and causes the rearrangement of an `activation segment'. Moreover, the mature N-terminus is repositioned to penetrate the CD moiety and is anchored to a buried `family-specific' glutamate. Overall, this mechanism of latency is reminiscent of that of the other three astacins with known zymogenic and mature structures, namely crayfish astacin, human meprin β and bacterial myroilysin, but each shows specific structural characteristics. Remarkably, myroilysin lacks the PP motif and employs a cysteine instead of the aspartate to block the catalytic metal.
- Proteolysis Laboratory, Department of Structural Biology, Molecular Biology Institute of Barcelona (IBMB), Higher Scientific Research Council (CSIC), Barcelona Science Park, Baldiri Reixac 15-21, Helix Building, 08028 Barcelona, Catalonia, Spain.
Organizational Affiliation: