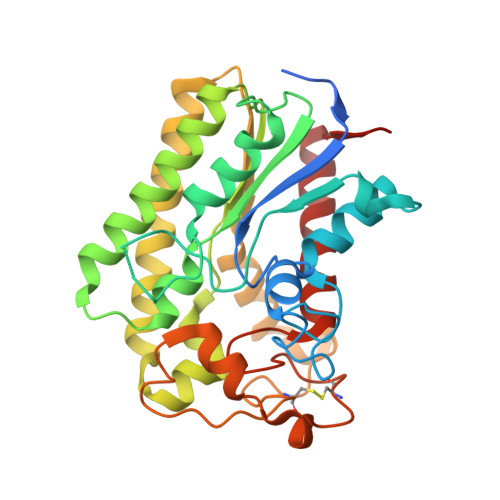
Structure-function relationships underpin disulfide loop cleavage-dependent activation of Legionella pneumophila lysophospholipase A PlaA.
Hiller, M., Diwo, M., Wamp, S., Gutsmann, T., Lang, C., Blankenfeldt, W., Flieger, A.(2024) Mol Microbiol 121: 497-512
- PubMed: 38130174
- DOI: https://doi.org/10.1111/mmi.15201
- Primary Citation of Related Structures:
8A24, 8A25, 8A26 - PubMed Abstract:
Legionella pneumophila, the causative agent of a life-threatening pneumonia, intracellularly replicates in a specialized compartment in lung macrophages, the Legionella-containing vacuole (LCV). Secreted proteins of the pathogen govern important steps in the intracellular life cycle including bacterial egress. Among these is the type II secreted PlaA which, together with PlaC and PlaD, belongs to the GDSL phospholipase family found in L. pneumophila. PlaA shows lysophospholipase A (LPLA) activity which increases after secretion and subsequent processing by the zinc metalloproteinase ProA within a disulfide loop. Activity of PlaA contributes to the destabilization of the LCV in the absence of the type IVB-secreted effector SdhA. We here present the 3D structure of PlaA which shows a typical α/β-hydrolase fold and reveals that the uncleaved disulfide loop forms a lid structure covering the catalytic triad S30/D278/H282. This leads to reduction of substrate access before activation; however, the catalytic site gets more accessible when the disulfide loop is processed. After structural modeling, a similar activation process is suggested for the GDSL hydrolase PlaC, but not for PlaD. Furthermore, the size of the PlaA substrate-binding site indicated preference toward phospholipids comprising ~16 carbon fatty acid residues which was verified by lipid hydrolysis, suggesting a molecular ruler mechanism. Indeed, mutational analysis changed the substrate profile with respect to fatty acid chain length. In conclusion, our analysis revealed the structural basis for the regulated activation and substrate preference of PlaA.
- Division of Enteropathogenic Bacteria and Legionella (FG11), Robert Koch Institute, Wernigerode, Germany.
Organizational Affiliation: