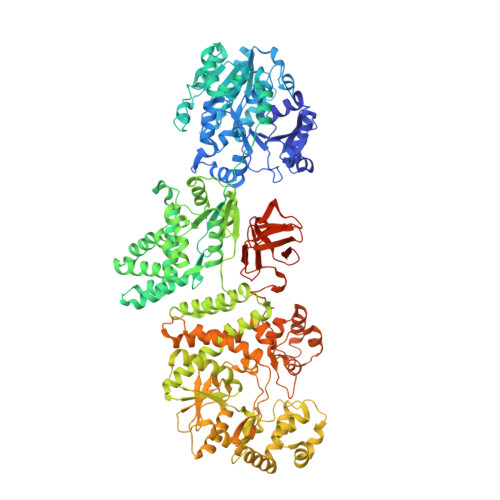
A multi-enzyme machine polymerizes the Haemophilus influenzae type b capsule.
Cifuente, J.O., Schulze, J., Bethe, A., Di Domenico, V., Litschko, C., Budde, I., Eidenberger, L., Thiesler, H., Ramon Roth, I., Berger, M., Claus, H., D'Angelo, C., Marina, A., Gerardy-Schahn, R., Schubert, M., Guerin, M.E., Fiebig, T.(2023) Nat Chem Biol 19: 865-877
- PubMed: 37277468
- DOI: https://doi.org/10.1038/s41589-023-01324-3
- Primary Citation of Related Structures:
8A0C, 8A0M - PubMed Abstract:
Bacterial capsules have critical roles in host-pathogen interactions. They provide a protective envelope against host recognition, leading to immune evasion and bacterial survival. Here we define the capsule biosynthesis pathway of Haemophilus influenzae serotype b (Hib), a Gram-negative bacterium that causes severe infections in infants and children. Reconstitution of this pathway enabled the fermentation-free production of Hib vaccine antigens starting from widely available precursors and detailed characterization of the enzymatic machinery. The X-ray crystal structure of the capsule polymerase Bcs3 reveals a multi-enzyme machine adopting a basket-like shape that creates a protected environment for the synthesis of the complex Hib polymer. This architecture is commonly exploited for surface glycan synthesis by both Gram-negative and Gram-positive pathogens. Supported by biochemical studies and comprehensive 2D nuclear magnetic resonance, our data explain how the ribofuranosyltransferase CriT, the phosphatase CrpP, the ribitol-phosphate transferase CroT and a polymer-binding domain function as a unique multi-enzyme assembly.
- Structural Glycobiology Laboratory, Biocruces Bizkaia Health Research Institute, Cruces University Hospital, Barakaldo, Spain.
Organizational Affiliation: