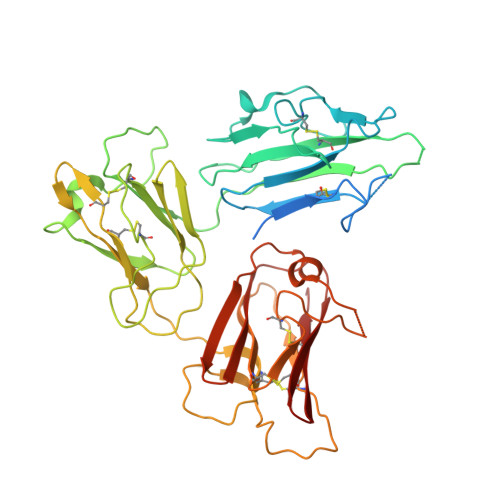
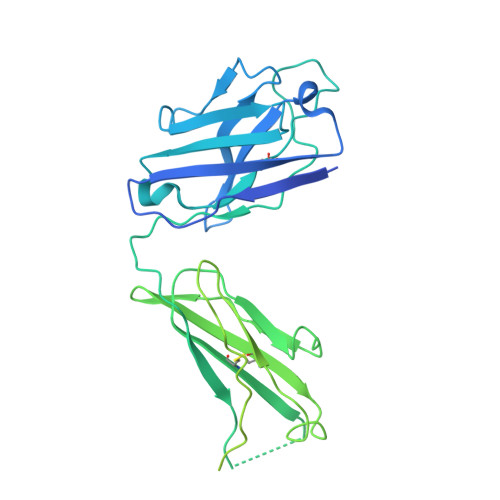
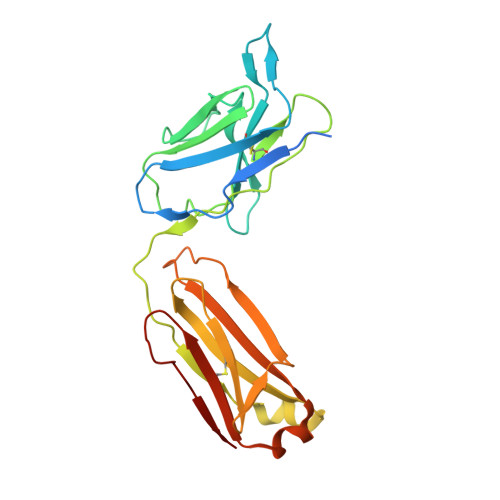
Structure of the malaria vaccine candidate Pfs48/45 and its recognition by transmission blocking antibodies.
Ko, K.T., Lennartz, F., Mekhaiel, D., Guloglu, B., Marini, A., Deuker, D.J., Long, C.A., Jore, M.M., Miura, K., Biswas, S., Higgins, M.K.(2022) Nat Commun 13: 5603-5603
- PubMed: 36153317
- DOI: https://doi.org/10.1038/s41467-022-33379-6
- Primary Citation of Related Structures:
7ZWF, 7ZWI, 7ZWM, 7ZXF, 7ZXG - PubMed Abstract:
An effective malaria vaccine remains a global health priority and vaccine immunogens which prevent transmission of the parasite will have important roles in multi-component vaccines. One of the most promising candidates for inclusion in a transmission-blocking malaria vaccine is the gamete surface protein Pfs48/45, which is essential for development of the parasite in the mosquito midgut. Indeed, antibodies which bind Pfs48/45 can prevent transmission if ingested with the parasite as part of the mosquito bloodmeal. Here we present the structure of full-length Pfs48/45, showing its three domains to form a dynamic, planar, triangular arrangement. We reveal where transmission-blocking and non-blocking antibodies bind on Pfs48/45. Finally, we demonstrate that antibodies which bind across this molecule can be transmission-blocking. These studies will guide the development of future Pfs48/45-based vaccine immunogens.
- Department of Biochemistry, South Parks Road, University of Oxford, Oxford, OX1 3QU, UK.
Organizational Affiliation: