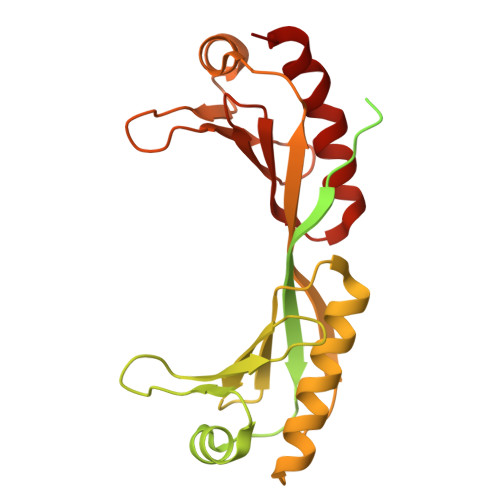
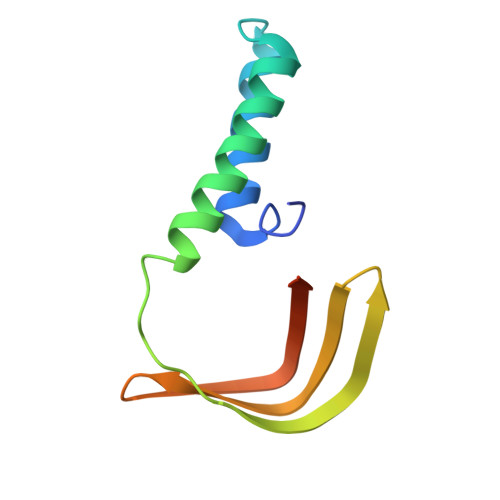
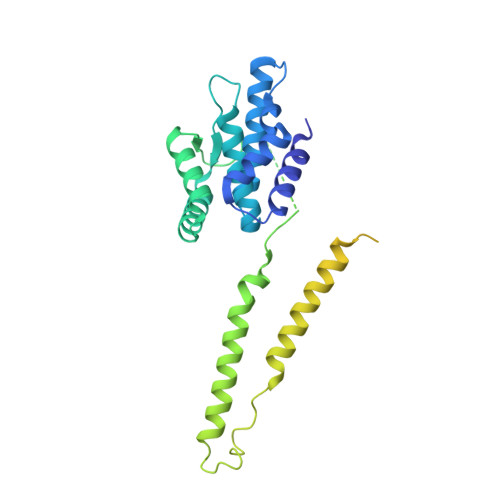
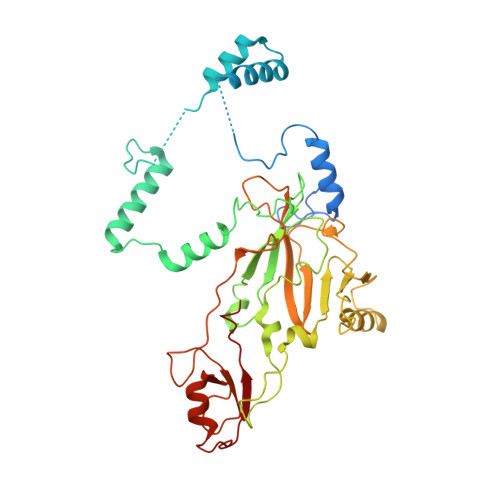
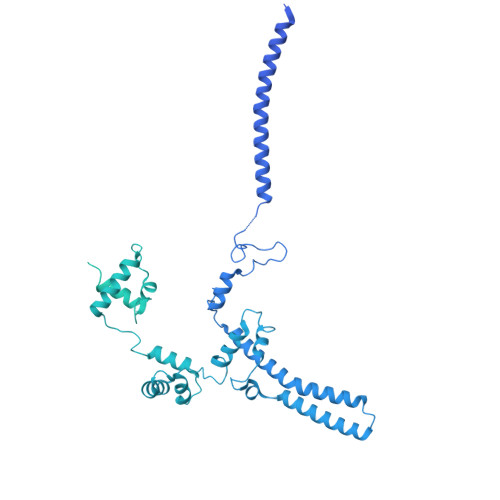
Structural basis of SNAPc-dependent snRNA transcription initiation by RNA polymerase II.
Rengachari, S., Schilbach, S., Kaliyappan, T., Gouge, J., Zumer, K., Schwarz, J., Urlaub, H., Dienemann, C., Vannini, A., Cramer, P.(2022) Nat Struct Mol Biol 29: 1159-1169
- PubMed: 36424526
- DOI: https://doi.org/10.1038/s41594-022-00857-w
- Primary Citation of Related Structures:
7ZWC, 7ZWD, 7ZX7, 7ZX8, 7ZXE - PubMed Abstract:
RNA polymerase II (Pol II) carries out transcription of both protein-coding and non-coding genes. Whereas Pol II initiation at protein-coding genes has been studied in detail, Pol II initiation at non-coding genes, such as small nuclear RNA (snRNA) genes, is less well understood at the structural level. Here, we study Pol II initiation at snRNA gene promoters and show that the snRNA-activating protein complex (SNAPc) enables DNA opening and transcription initiation independent of TFIIE and TFIIH in vitro. We then resolve cryo-EM structures of the SNAPc-containing Pol IIpre-initiation complex (PIC) assembled on U1 and U5 snRNA promoters. The core of SNAPc binds two turns of DNA and recognizes the snRNA promoter-specific proximal sequence element (PSE), located upstream of the TATA box-binding protein TBP. Two extensions of SNAPc, called wing-1 and wing-2, bind TFIIA and TFIIB, respectively, explaining how SNAPc directs Pol II to snRNA promoters. Comparison of structures of closed and open promoter complexes elucidates TFIIH-independent DNA opening. These results provide the structural basis of Pol II initiation at non-coding RNA gene promoters.
- Department of Molecular Biology, Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany.
Organizational Affiliation: