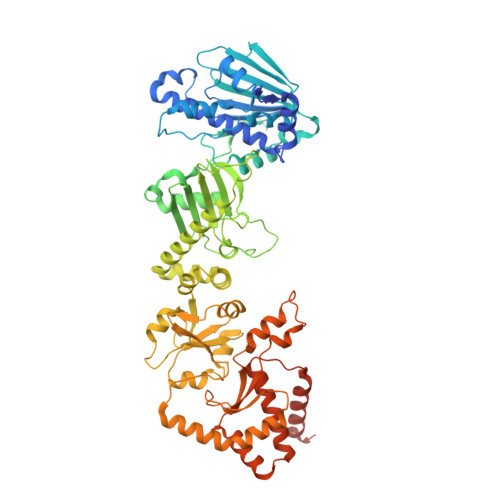
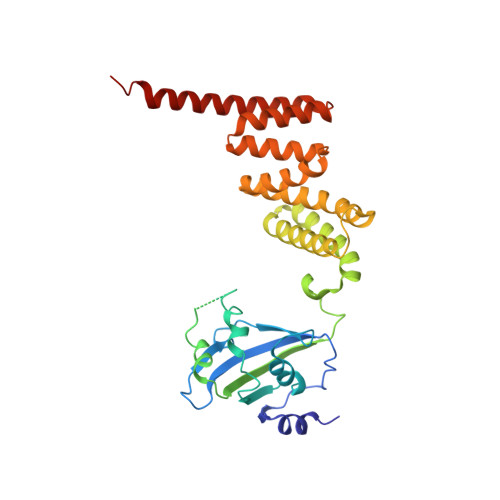
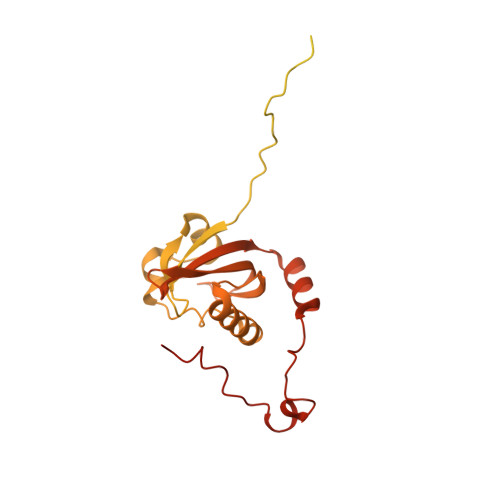
Cryo-EM structure of the agonist-bound Hsp90-XAP2-AHR cytosolic complex.
Gruszczyk, J., Grandvuillemin, L., Lai-Kee-Him, J., Paloni, M., Savva, C.G., Germain, P., Grimaldi, M., Boulahtouf, A., Kwong, H.S., Bous, J., Ancelin, A., Bechara, C., Barducci, A., Balaguer, P., Bourguet, W.(2022) Nat Commun 13: 7010-7010
- PubMed: 36385050
- DOI: https://doi.org/10.1038/s41467-022-34773-w
- Primary Citation of Related Structures:
7ZUB - PubMed Abstract:
The aryl hydrocarbon receptor (AHR) is a ligand-dependent transcription factor that mediates a broad spectrum of (patho)physiological processes in response to numerous substances including pollutants, natural products and metabolites. However, the scarcity of structural data precludes understanding of how AHR is activated by such diverse compounds. Our 2.85 Å structure of the human indirubin-bound AHR complex with the chaperone Hsp90 and the co-chaperone XAP2, reported herein, reveals a closed conformation Hsp90 dimer with AHR threaded through its lumen and XAP2 serving as a brace. Importantly, we disclose the long-awaited structure of the AHR PAS-B domain revealing a unique organisation of the ligand-binding pocket and the structural determinants of ligand-binding specificity and promiscuity of the receptor. By providing structural details of the molecular initiating event leading to AHR activation, our study rationalises almost forty years of biochemical data and provides a framework for future mechanistic studies and structure-guided drug design.
- CBS (Centre de Biologie Structurale), Univ Montpellier, CNRS, Inserm, Montpellier, France. jakub.gruszczyk@cbs.cnrs.fr.
Organizational Affiliation: