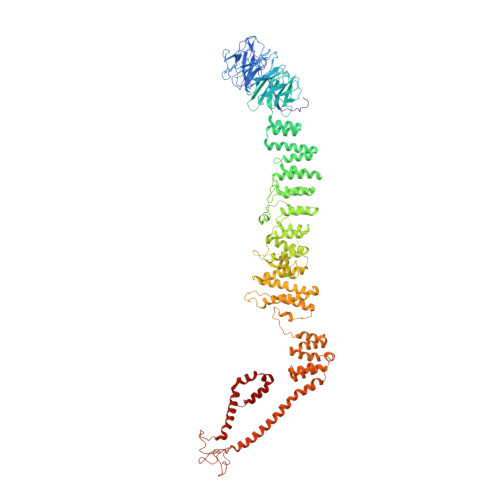
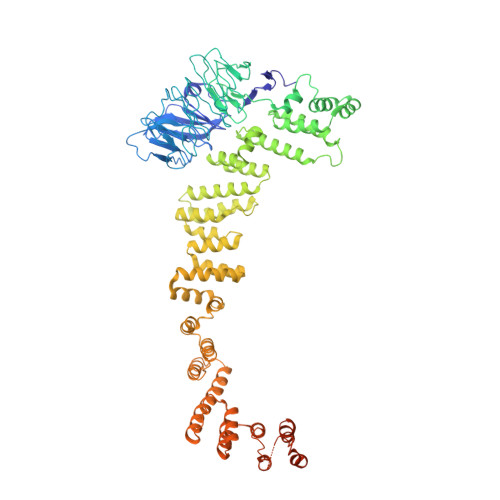
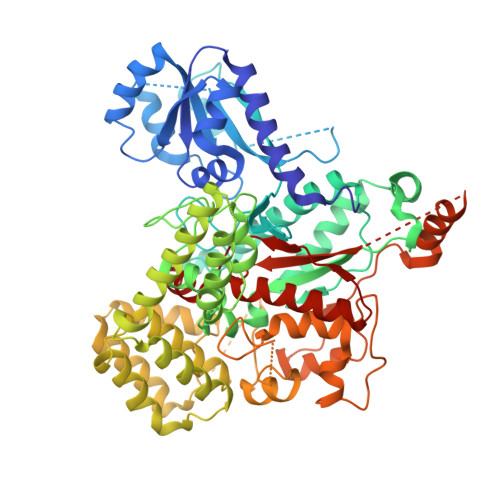
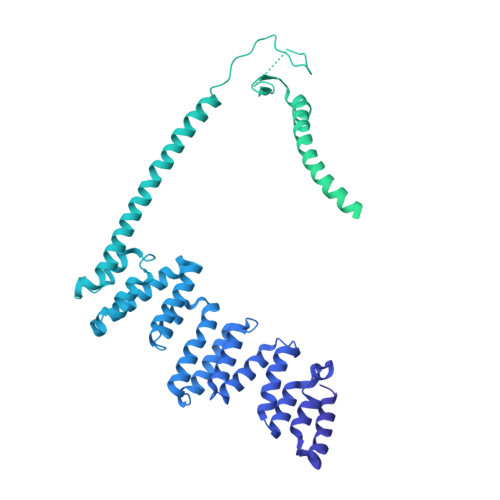
Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery.
Shvarev, D., Schoppe, J., Konig, C., Perz, A., Fullbrunn, N., Kiontke, S., Langemeyer, L., Januliene, D., Schnelle, K., Kummel, D., Frohlich, F., Moeller, A., Ungermann, C.(2022) Elife 11
- PubMed: 36098503
- DOI: https://doi.org/10.7554/eLife.80901
- Primary Citation of Related Structures:
7ZTY, 7ZU0 - PubMed Abstract:
Lysosomes are essential for cellular recycling, nutrient signaling, autophagy, and pathogenic bacteria and viruses invasion. Lysosomal fusion is fundamental to cell survival and requires HOPS, a conserved heterohexameric tethering complex. On the membranes to be fused, HOPS binds small membrane-associated GTPases and assembles SNAREs for fusion, but how the complex fulfills its function remained speculative. Here, we used cryo-electron microscopy to reveal the structure of HOPS. Unlike previously reported, significant flexibility of HOPS is confined to its extremities, where GTPase binding occurs. The SNARE-binding module is firmly attached to the core, therefore, ideally positioned between the membranes to catalyze fusion. Our data suggest a model for how HOPS fulfills its dual functionality of tethering and fusion and indicate why it is an essential part of the membrane fusion machinery.
- Department of Biology/Chemistry, Structural Biology section, Osnabrück University, Osnabrück, Germany.
Organizational Affiliation: