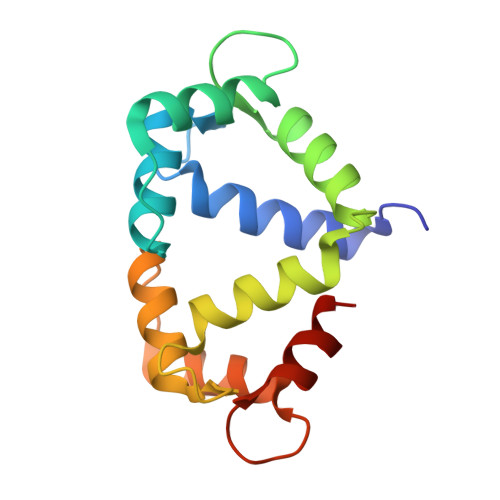
Calmodulin variant E140G associated with long QT syndrome impairs CaMKII delta autophosphorylation and L-type calcium channel inactivation.
Prakash, O., Gupta, N., Milburn, A., McCormick, L., Deugi, V., Fisch, P., Wyles, J., Thomas, N.L., Antonyuk, S., Dart, C., Helassa, N.(2022) J Biological Chem 299: 102777-102777
- PubMed: 36496072
- DOI: https://doi.org/10.1016/j.jbc.2022.102777
- Primary Citation of Related Structures:
7ZRP, 7ZRQ - PubMed Abstract:
Long QT syndrome (LQTS) is a human inherited heart condition that can cause life-threatening arrhythmia including sudden cardiac death. Mutations in the ubiquitous Ca 2+ -sensing protein calmodulin (CaM) are associated with LQTS, but the molecular mechanism by which these mutations lead to irregular heartbeats is not fully understood. Here, we use a multidisciplinary approach including protein biophysics, structural biology, confocal imaging, and patch-clamp electrophysiology to determine the effect of the disease-associated CaM mutation E140G on CaM structure and function. We present novel data showing that mutant-regulated CaMKIIδ kinase activity is impaired with a significant reduction in enzyme autophosphorylation rate. We report the first high-resolution crystal structure of a LQTS-associated CaM variant in complex with the CaMKIIδ peptide, which shows significant structural differences, compared to the WT complex. Furthermore, we demonstrate that the E140G mutation significantly disrupted Ca v 1.2 Ca 2+ /CaM-dependent inactivation, while cardiac ryanodine receptor (RyR2) activity remained unaffected. In addition, we show that the LQTS-associated mutation alters CaM's Ca 2+ -binding characteristics, secondary structure content, and interaction with key partners involved in excitation-contraction coupling (CaMKIIδ, Ca v 1.2, RyR2). In conclusion, LQTS-associated CaM mutation E140G severely impacts the structure-function relationship of CaM and its regulation of CaMKIIδ and Ca v 1.2. This provides a crucial insight into the molecular factors contributing to CaM-mediated arrhythmias with a central role for CaMKIIδ.
- Liverpool Centre for Cardiovascular Science, Department of Cardiovascular and Metabolic Medicine, Institute of Life Course and Medical Sciences, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, United Kingdom.
Organizational Affiliation: