Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Rotheneder, M., Stakyte, K., van de Logt, E., Bartho, J.D., Lammens, K., Fan, Y., Alt, A., Kessler, B., Jung, C., Roos, W.P., Steigenberger, B., Hopfner, K.P.(2023) Mol Cell 83: 167-185.e9
- PubMed: 36577401
- DOI: https://doi.org/10.1016/j.molcel.2022.12.003
- Primary Citation of Related Structures:
7ZQY, 7ZR1, 8BAH - PubMed Abstract:
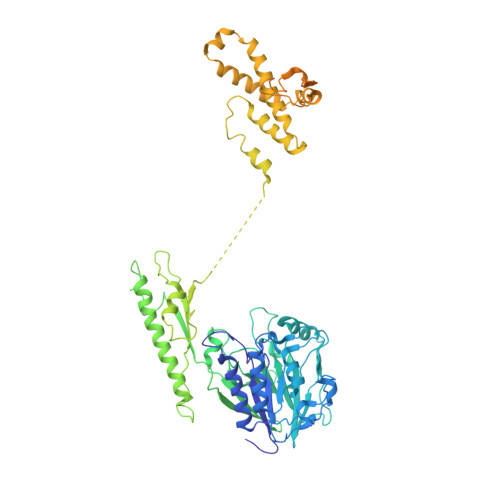
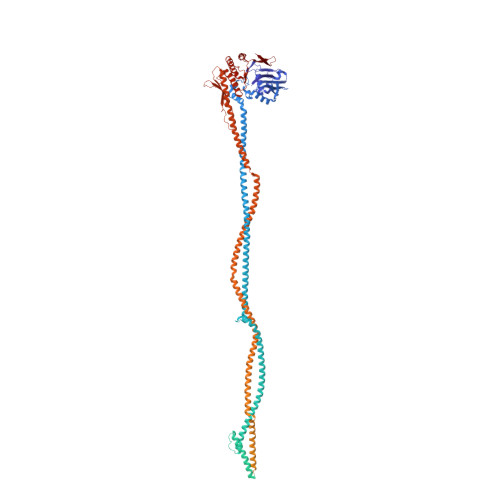
The DNA double-strand break repair complex Mre11-Rad50-Nbs1 (MRN) detects and nucleolytically processes DNA ends, activates the ATM kinase, and tethers DNA at break sites. How MRN can act both as nuclease and scaffold protein is not well understood. The cryo-EM structure of MRN from Chaetomium thermophilum reveals a 2:2:1 complex with a single Nbs1 wrapping around the autoinhibited Mre11 nuclease dimer. MRN has two DNA-binding modes, one ATP-dependent mode for loading onto DNA ends and one ATP-independent mode through Mre11's C terminus, suggesting how it may interact with DSBs and intact DNA. MRNs two 60-nm-long coiled-coil domains form a linear rod structure, the apex of which is assembled by the two joined zinc-hook motifs. Apices from two MRN complexes can further dimerize, forming 120-nm spanning MRN-MRN structures. Our results illustrate the architecture of MRN and suggest how it mechanistically integrates catalytic and tethering functions.
- Gene Center, Department of Biochemistry, Ludwig Maximilians Universität, Munich, Germany.
Organizational Affiliation: