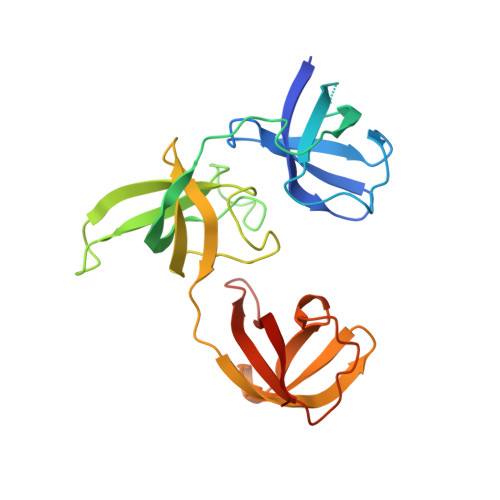
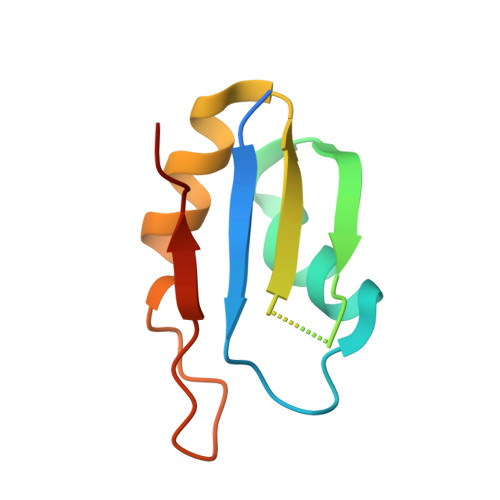
Upstream of N-Ras C-terminal cold shock domains mediate poly(A) specificity in a novel RNA recognition mode and bind poly(A) binding protein.
Hollmann, N.M., Jagtap, P.K.A., Linse, J.B., Ullmann, P., Payr, M., Murciano, B., Simon, B., Hub, J.S., Hennig, J.(2023) Nucleic Acids Res 51: 1895-1913
- PubMed: 36688322
- DOI: https://doi.org/10.1093/nar/gkac1277
- Primary Citation of Related Structures:
7ZHH, 7ZHR - PubMed Abstract:
RNA binding proteins (RBPs) often engage multiple RNA binding domains (RBDs) to increase target specificity and affinity. However, the complexity of target recognition of multiple RBDs remains largely unexplored. Here we use Upstream of N-Ras (Unr), a multidomain RBP, to demonstrate how multiple RBDs orchestrate target specificity. A crystal structure of the three C-terminal RNA binding cold-shock domains (CSD) of Unr bound to a poly(A) sequence exemplifies how recognition goes beyond the classical ππ-stacking in CSDs. Further structural studies reveal several interaction surfaces between the N-terminal and C-terminal part of Unr with the poly(A)-binding protein (pAbp). All interactions are validated by mutational analyses and the high-resolution structures presented here will guide further studies to understand how both proteins act together in cellular processes.
- Structural and Computational Biology Unit, EMBL Heidelberg, Meyerhofstraße 1, 69117 Heidelberg, Germany.
Organizational Affiliation: