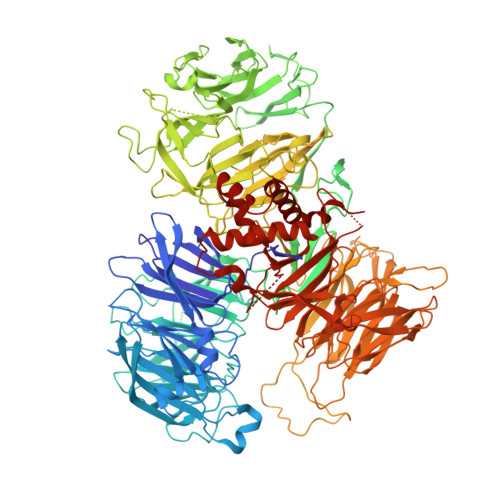
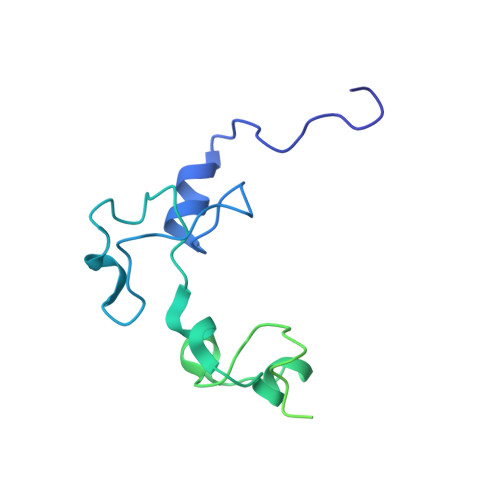
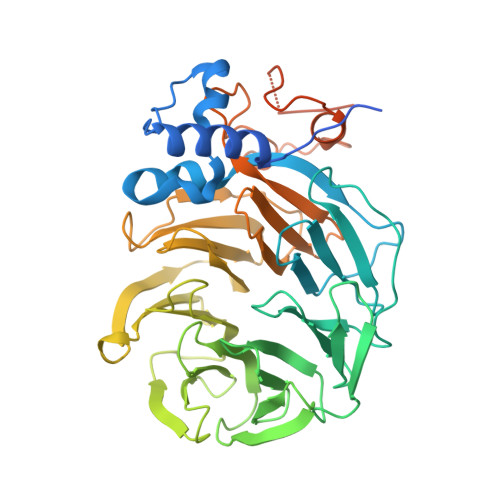
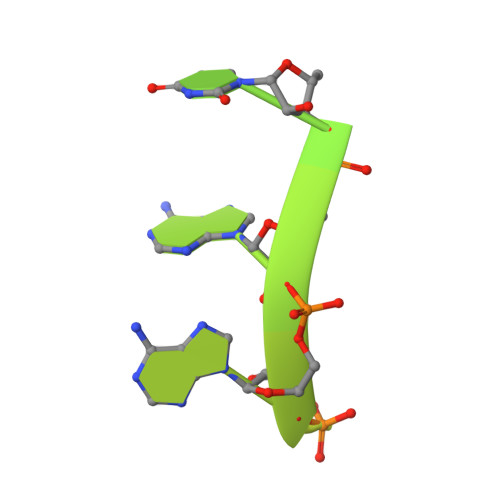
Mpe1 senses the binding of pre-mRNA and controls 3' end processing by CPF.
Rodriguez-Molina, J.B., O'Reilly, F.J., Fagarasan, H., Sheekey, E., Maslen, S., Skehel, J.M., Rappsilber, J., Passmore, L.A.(2022) Mol Cell 82: 2490
- PubMed: 35584695
- DOI: https://doi.org/10.1016/j.molcel.2022.04.021
- Primary Citation of Related Structures:
7ZGP, 7ZGQ, 7ZGR - PubMed Abstract:
Most eukaryotic messenger RNAs (mRNAs) are processed at their 3' end by the cleavage and polyadenylation specificity factor (CPF/CPSF). CPF mediates the endonucleolytic cleavage of the pre-mRNA and addition of a polyadenosine (poly(A)) tail, which together define the 3' end of the mature transcript. The activation of CPF is highly regulated to maintain the fidelity of RNA processing. Here, using cryo-EM of yeast CPF, we show that the Mpe1 subunit directly contacts the polyadenylation signal sequence in nascent pre-mRNA. The region of Mpe1 that contacts RNA also promotes the activation of CPF endonuclease activity and controls polyadenylation. The Cft2 subunit of CPF antagonizes the RNA-stabilized configuration of Mpe1. In vivo, the depletion or mutation of Mpe1 leads to widespread defects in transcription termination by RNA polymerase II, resulting in transcription interference on neighboring genes. Together, our data suggest that Mpe1 plays a major role in accurate 3' end processing, activating CPF, and ensuring timely transcription termination.
- MRC Laboratory of Molecular Biology, Cambridge CB2 0QH, UK.
Organizational Affiliation: