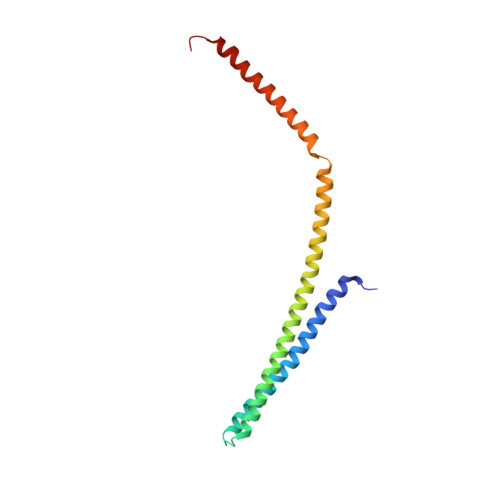
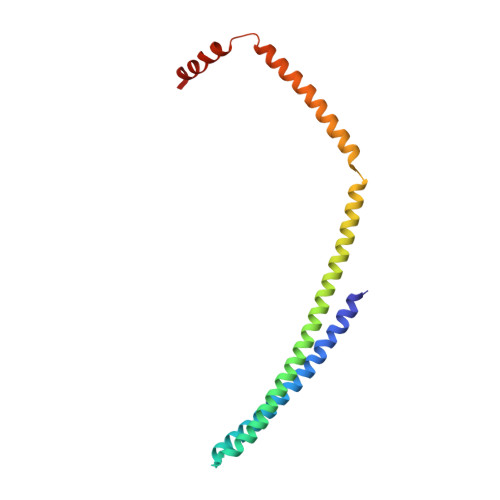
Structural basis of CHMP2A-CHMP3 ESCRT-III polymer assembly and membrane cleavage.
Azad, K., Guilligay, D., Boscheron, C., Maity, S., De Franceschi, N., Sulbaran, G., Effantin, G., Wang, H., Kleman, J.P., Bassereau, P., Schoehn, G., Roos, W.H., Desfosses, A., Weissenhorn, W.(2023) Nat Struct Mol Biol 30: 81-90
- PubMed: 36604498
- DOI: https://doi.org/10.1038/s41594-022-00867-8
- Primary Citation of Related Structures:
7ZCG, 7ZCH - PubMed Abstract:
The endosomal sorting complex required for transport (ESCRT) is a highly conserved protein machinery that drives a divers set of physiological and pathological membrane remodeling processes. However, the structural basis of ESCRT-III polymers stabilizing, constricting and cleaving negatively curved membranes is yet unknown. Here we present cryo-EM structures of membrane-coated CHMP2A-CHMP3 filaments from Homo sapiens of two different diameters at 3.3 and 3.6 Å resolution. The structures reveal helical filaments assembled by CHMP2A-CHMP3 heterodimers in the open ESCRT-III conformation, which generates a partially positive charged membrane interaction surface, positions short N-terminal motifs for membrane interaction and the C-terminal VPS4 target sequence toward the tube interior. Inter-filament interactions are electrostatic, which may facilitate filament sliding upon VPS4-mediated polymer remodeling. Fluorescence microscopy as well as high-speed atomic force microscopy imaging corroborate that VPS4 can constrict and cleave CHMP2A-CHMP3 membrane tubes. We therefore conclude that CHMP2A-CHMP3-VPS4 act as a minimal membrane fission machinery.
- Institute of Structural Biology (IBS), University Grenoble Alpes, CEA, CNRS, Grenoble, France.
Organizational Affiliation: