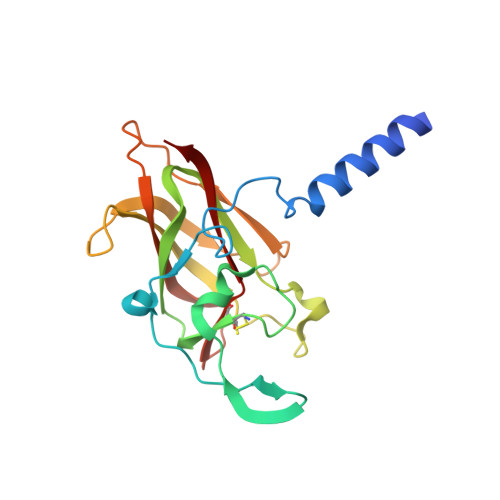
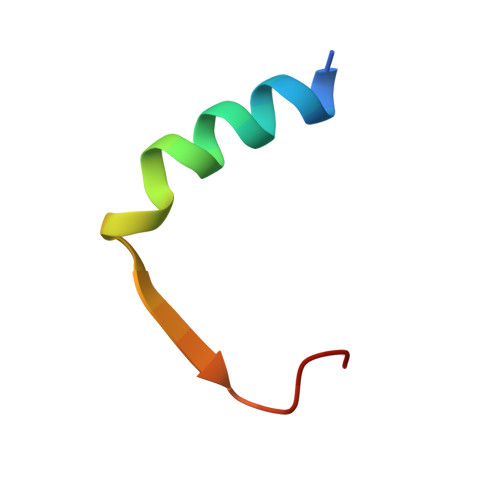
The crystal structure of SUN1-KASH6 reveals an asymmetric LINC complex architecture compatible with nuclear membrane insertion.
Gurusaran, M., Erlandsen, B.S., Davies, O.R.(2024) Commun Biol 7: 138-138
- PubMed: 38291267
- DOI: https://doi.org/10.1038/s42003-024-05794-6
- Primary Citation of Related Structures:
7Z8Y, 8B46, 8B5X - PubMed Abstract:
The LINC complex transmits cytoskeletal forces into the nucleus to control the structure and movement of nuclear contents. It is formed of nuclear SUN and cytoplasmic KASH proteins, which interact within the nuclear lumen, immediately below the outer nuclear membrane. However, the symmetrical location of KASH molecules within SUN-KASH complexes in previous crystal structures has been difficult to reconcile with the steric requirements for insertion of their immediately upstream transmembrane helices into the outer nuclear membrane. Here, we report the crystal structure of the SUN-KASH complex between SUN1 and JAW1/LRMP (KASH6) in an asymmetric 9:6 configuration. This intertwined assembly involves two distinct KASH conformations such that all six KASH molecules emerge on the same molecular surface. Hence, they are ideally positioned for insertion of upstream sequences into the outer nuclear membrane. Thus, we report a SUN-KASH complex architecture that appears to be directly compatible with its biological role.
- Wellcome Centre for Cell Biology, Institute of Cell Biology, University of Edinburgh, Michael Swann Building, Max Born Crescent, Edinburgh, EH9 3BF, UK.
Organizational Affiliation: