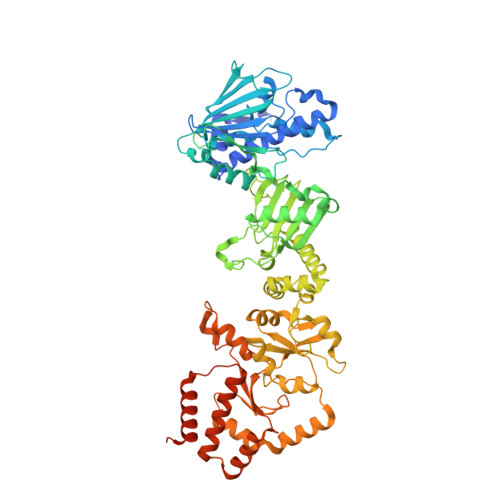
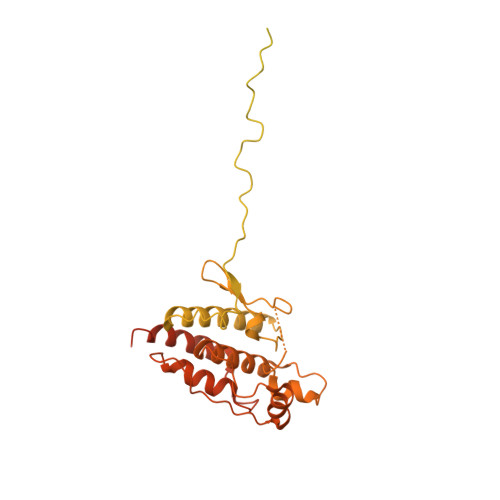
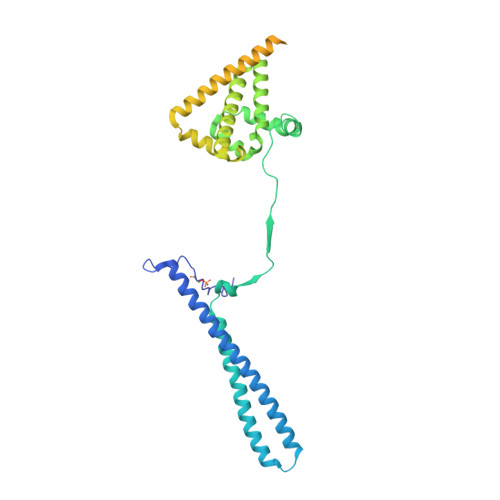
Structure of the RAF1-HSP90-CDC37 complex reveals the basis of RAF1 regulation.
Garcia-Alonso, S., Mesa, P., Ovejero, L.P., Aizpurua, G., Lechuga, C.G., Zarzuela, E., Santiveri, C.M., Sanclemente, M., Munoz, J., Musteanu, M., Campos-Olivas, R., Martinez-Torrecuadrada, J., Barbacid, M., Montoya, G.(2022) Mol Cell 82: 3438-3452.e8
- PubMed: 36055235
- DOI: https://doi.org/10.1016/j.molcel.2022.08.012
- Primary Citation of Related Structures:
7Z37, 7Z38 - PubMed Abstract:
RAF kinases are RAS-activated enzymes that initiate signaling through the MAPK cascade to control cellular proliferation, differentiation, and survival. Here, we describe the structure of the full-length RAF1 protein in complex with HSP90 and CDC37 obtained by cryoelectron microscopy. The reconstruction reveals a RAF1 kinase with an unfolded N-lobe separated from its C-lobe. The hydrophobic core of the N-lobe is trapped in the HSP90 dimer, while CDC37 wraps around the chaperone and interacts with the N- and C-lobes of the kinase. The structure indicates how CDC37 can discriminate between the different members of the RAF family. Our structural analysis also reveals that the folded RAF1 assembles with 14-3-3 dimers, suggesting that after folding RAF1 follows a similar activation as B-RAF. Finally, disruption of the interaction between CDC37 and the DFG segment of RAF1 unveils potential vulnerabilities in attempting the pharmacological degradation of RAF1 for therapeutic purposes.
- Experimental Oncology Group, Molecular Oncology Programme, Centro Nacional de Investigaciones Oncológicas (CNIO), Madrid 28029, Spain; Centro de Investigación Biomédica en Red de Cáncer (CIBERONC), Instituto de Salud Carlos III, 28029 Madrid, Spain.
Organizational Affiliation: